Figure 5.
Exploring myofibroblast states at D1 and epithelial-mesenchymal regulatory modules guiding alveolar septation
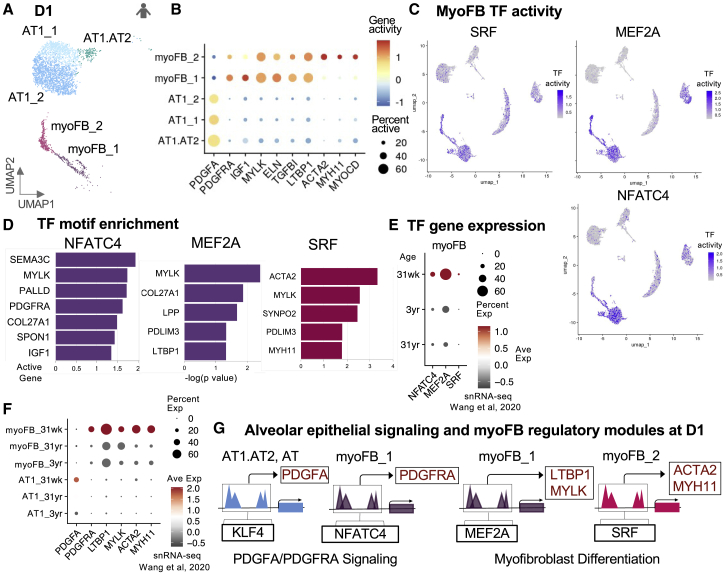
(A) UMAP of annotated D1 AT1.AT2, AT1 and myoFB clusters.
(B) Dot plot of log-normalized gene activity scores for PDGFA and select myoFB DA genes.
(C) UMAP from Figure S3A re-colored with TF activity levels for key myoFB TFs.
(D) Bar plots of -log10(p values) for select active genes in myoFB_1 or myoFB_2 cells with significant NFATC, MEF2, and SRF motif enrichment.
(E) Dot plot of TF gene expression in Wang et al. snRNA-seq by myoFBs grouped by BRINDL donor age, ∼30 weeks gestational age, 3, and 31 years old.
(F) Dot plot of active gene expression in Wang et al. snRNA-seq grouped by donor age.
(G) Proposed regulatory modules for D1 PDGFA:PDGFRA signaling and myoFB differentiation. All marker genes in rectangles contained peaks enriched in TF motifs and are expressed in AT1 or myoFB clusters in Wang et al. sn RNA-seq at ∼30 week D1.
See also Table S4.