Extended Data Fig. 5. Signal to noise in virus mixtures containing SARS-CoV-2 full spike and HIV env.
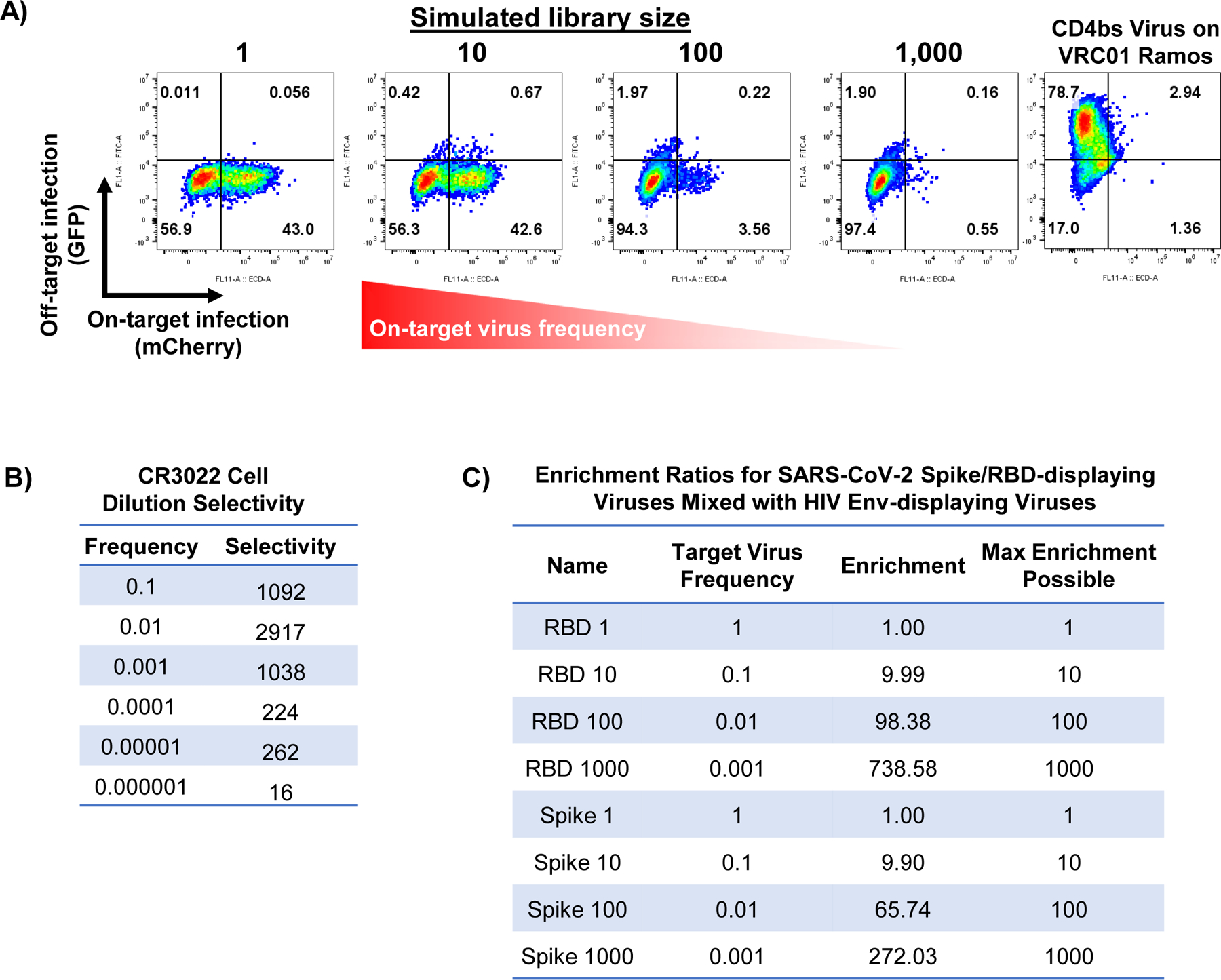
A, Infection of CR3022 BCR-expressing Ramos cells with mixtures of SARS-CoV-2 prefusion stabilized spike (on-target, expressing mCherry) and HIV env (off-target, expressing GFP) viruses; variant library size indicates the ratio of off-target to on-target virus present. Infection of VRC01-expressing Ramos cells with HIV env hybrid pseudotyped viruses (right) demonstrates that the viral particles remain functional. B, Quantification of selectivity ratios for the SARS-CoV-2 RBD infections of CR3022 Ramos presented in Figure 5C. Selectivity ratio is calculated as the proportion of on-target cells infected divided by the proportion of off-target cells infected times the frequency of on-target cells (see Methods). C, Quantification of on-target enrichment for mixtures of HIV env and either SARS-CoV-2 RBD (presented in Fig. 5D) or full spike (presented in Supplementary Fig. 5A). Enrichment is calculated by fraction of cells expressing on-target signal divided by fraction of cells expressing off-target signal times on-target virus frequency.
