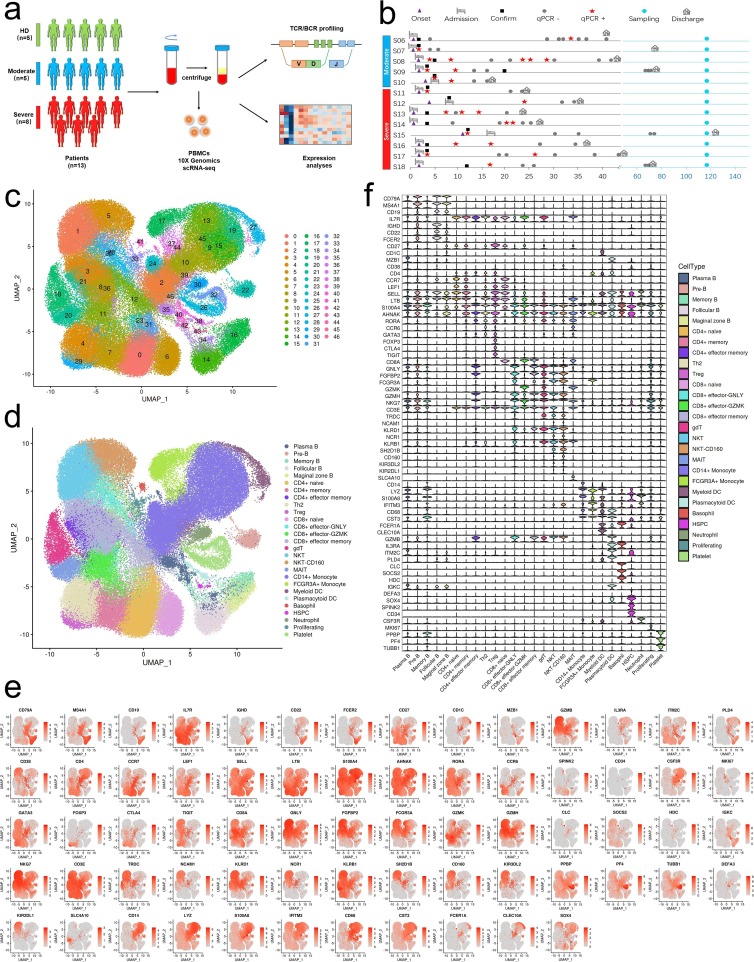
Fig. 1.
Study design and single-cell transcriptional profiling of PBMCs from HDs and convalescent COVID-19 patients. a, Study design and single-cell transcriptional profiling of PBMCs from HDs and convalescent COVID-19 patients. A schematic showing the overall study design. The scRNA-seq was applied to PBMCs across three conditions and the output data were used for TCR and BCR profiling and expression analyses. b, Timeline of the course of disease for 13 patients infected with SARS-CoV-2 and enrolled in our study. qPCR indicates PCR testing for SARS-CoV-2 nucleic acids. qPCR positive indicates nasopharyngeal swab samples that were positive for SARS-CoV-2 nucleic acids. Healthy donors are colored in green, convalescent patients with moderate COVID-19 are colored in blue and convalescent severe patients are colored in reds. The color bars on the left represent conditions with the same color as in a. c, Quality control and cell populations identified. A total of 17 PBMC samples with HDs (n = 4), and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8), were sequenced and obtained a total of 122,726 high-quality single cells. After quality control, 117,102 cells were obtained from 122,726 of PBMCs, 47 clusters of cells were identified by UMAP. Each dot corresponds to a single cell, colored according to cell type. d, 27 Cellular populations identified. The 47 clusters of cells in Fig. 1c were identified further by cell types. The UMAP projection of 117,102 single cells from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8), showing 27 clusters with the respective labels. The 27 clusters were identified and showed with UMAP. Each dot corresponds to a single cell, colored according to cell type. e, Canonical cell markers were used to label clusters by cell identity as represented in the UMAP plot. Data are colored according to expression levels and the legend is labeled in log scale. f, Canonical cell markers were used to label clusters by cell identity in 27 clusters as represented in the violin plot. Y-axis represents log2 (normalized count + 1).