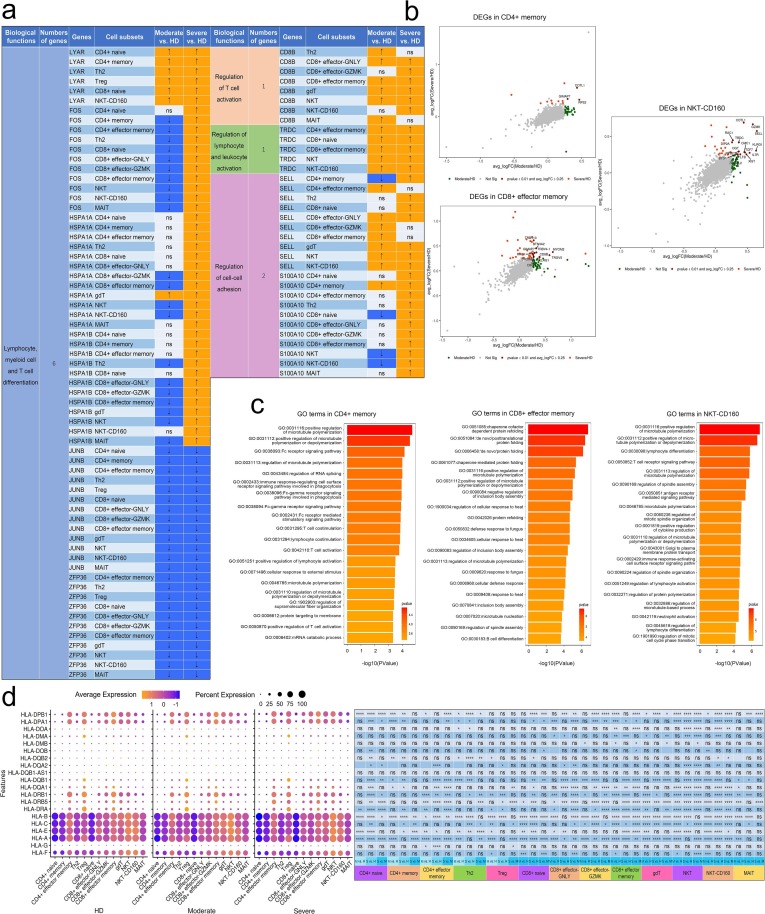
Fig. 5.
DEGs in T cells in individuals across three conditions. a, Expression pattern of DEGs relevant to specific biological functions in regulating T cell immunity during convalescence of COVID-19. DEGs relevant to specific biological functions in regulating innate immunity were identified from Fig. 4d and expression pattern of these DEGs in cell subsets were shown. Up arrow was correspondence to upregulation and down arrow was correspondence to downregulation. ns represented as no significance. b, Scatter-plot showing DEGs in T cell subsets (CD4 + Memory, CD8 + Effector memory and NKT-CD-160) derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) calculated by Markers (Seurat v3.2.0, default parameters) function. Each red dot denotes an individual gene with P < 0.01 and average log2 fold change greater than 0.25. The plots with the gene symbols are the genes which are significantly differentially expressed both when convalescent patients with moderate COVID-19 compared with HDs groups and convalescent patients with severe COVID-19 compared with HDs. Orange dots indicate the genes which are only significantly differentially expressed when convalescent patients with severe COVID-19 compared with HDs. Green dots indicate the genes which are only significantly differentially expressed when convalescent patients with moderate COVID-19 compared with HDs. c, Gene enrichment analyses of the DEGs in Fig. 4d in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown. d, Dot plot depicts percent expression and average expression of all detected HLA genes in T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The size of the dot represents the expression percentage of T cell clusters, and data color represents the expression levels after log scale. The right panel shows differential expression significance between the three groups with unpaired two-sided t-test.