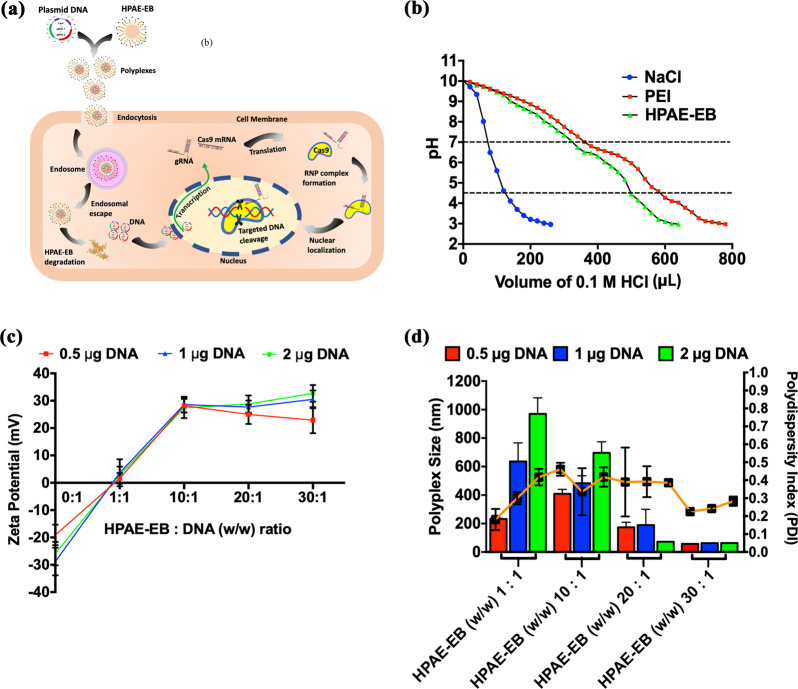
Fig. 3. Transfection process and physical characterisation of HPAE-EB polymer alone and complexed to CRISPR-C7 plasmid.
a Non-viral transfection process using HPAE-EB and CRISPR-C7 plasmid DNA to mediate gene editing. b Endosomal buffering capacity of the HPAE-EB polymer. Acid–base titration curve of HPAE-EB, PEI (25 kDa) and NaCl across a wide pH range was performed. PEI (25 kDa) was used as a commercial standard for comparison and NaCl was used as a negative control. With addition of HCl, NaCl shows sharp decline in pH, whereas PEI and HPAE-EB exhibit gradual decrease owing to higher proton buffering capacity (n = 3). c Zeta potential of HPAE-EB polyplexes with CRISPR-C7 plasmid DNA, complexed at different w/w ratios. DNA alone (0:1 w/w ratio) was used to demonstrate the negative charge of DNA, and from w/w ratios of 10:1 and higher, polyplex surface charge is positive for all DNA quantities. d Hydrodynamic size and PDI distribution of the HPAE-EB polymer and CRISPR-C7 plasmid DNA polyplexes at varying DNA quantities and polymer:DNA w/w ratios. As polymer:DNA ratio is increased, there is a sharp decrease in polyplex size across all DNA quantities. PDI distribution is heterogenous throughout all conditions tested. Data are presented as mean ± SD of three replicates from three independent experiments (n = 3).