Figure 2.
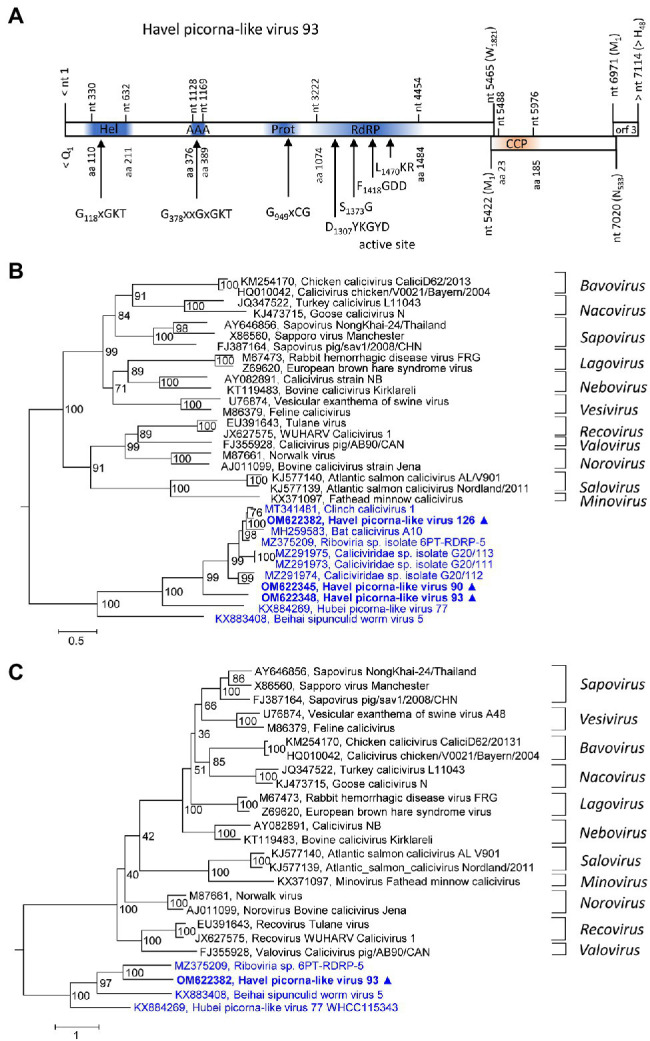
Analysis of new caliciviruses. (A) The genome organization of Havel picorna-like virus 93 resembles the calicivirus genome. ORF1 shows conserved elements of helicase (Hel), proteinase (Prot), and polymerase (RdRP) domains, whereas ORF2 has similarity to the calicivirus capsid protein (CCP) family of caliciviruses. Conserved domains and sequence motifs were identified by searching the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Phylogenetic analyses of the VP1 (capsid protein) gene region (B) and the proteinase/polymerase gene region (C) of calicivirus prototype strains, unassigned caliciviruses, and Havel picorna-like viruses 93, −175, and −176 are shown. The trees were inferred with IQ-Tree 2, optimal substitution model: TVM + F + R4 (B) and TVMe+R4 (C). Numbers at nodes present bootstrap values obtained after 50,000 ultrafast bootstrap replications. The scale indicates substitutions per site. Presented are GenBank acc. nos. and virus names. The respective genera are indicated to the right. Unassigned viruses are printed in blue. A triangle (▲) indicates the viruses of the present study. AAA, AAA protein with homology to ATPase family associated with various cellular functions, Hel, helicase, Prot, proteinase; RdRP, polymerase; and CCP, capsid protein with similarity to the calicivirus capsid protein.
