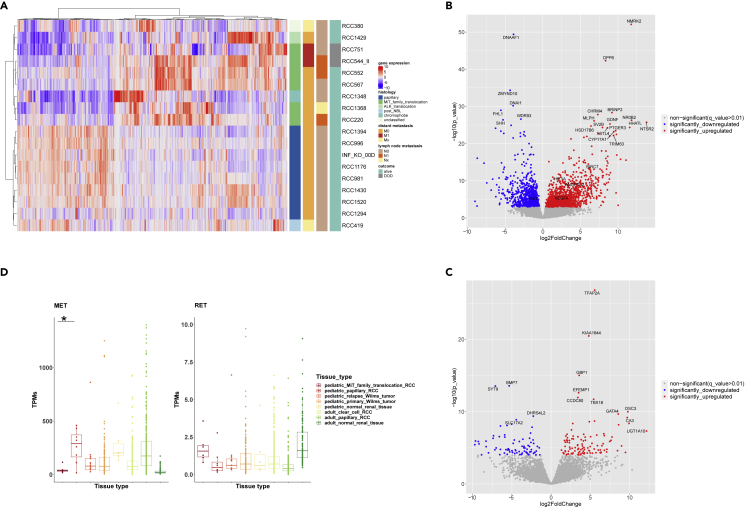
Figure 4.
Gene expression analysis (RNA-seq data) of pediatric RCC (pRCC; n = 18). Among the 18/25 pRCC cases with available RNA, 6 MiT-pRCC cases (4 with ASPSCR1-TFE3 fusion, 1 with TFE3-SFPQ fusion, and 1 with TFEB-CLTC fusion), 8 papillary pRCC (6/8 histologically papillary type 1), one chromophobe, one ALK-rearranged, one post-NB, and one FH-deficient RCC were included
(A) Unsupervised hierarchical clustering of pRCC samples based on the top 1,000 most differentially expressed genes (DEGs).
(B) Volcano-plot illustration of the significantly DEGs in MiT-pRCC versus papillary pRCC.
(C) Volcano-plot illustration of the significantly DEGs in metastatic fatal MiT-pRCC versus nonmetastatic surviving MiT-pRCC.
(D) MET and RET gene expression in MiT-pRCC and papillary pRCC (ppRCC) compared with Wilms tumor relapses (INFORM), primary Wilms tumors, and normal pediatric kidney (TARGET) as well as adult clear cell RCC, adult papillary RCC, and adult normal kidney (TCGA). MET gene expression significantly higher in ppRCC versus MiT-pRCC, log2FC = 1.6, p < 0.0001, q < 0.01 (Wald test). RET gene expression significantly higher in MiT-pRCC versus ppRCC, log2FC = 1.15, p = 0.013, q = 0.056 (Wald test).