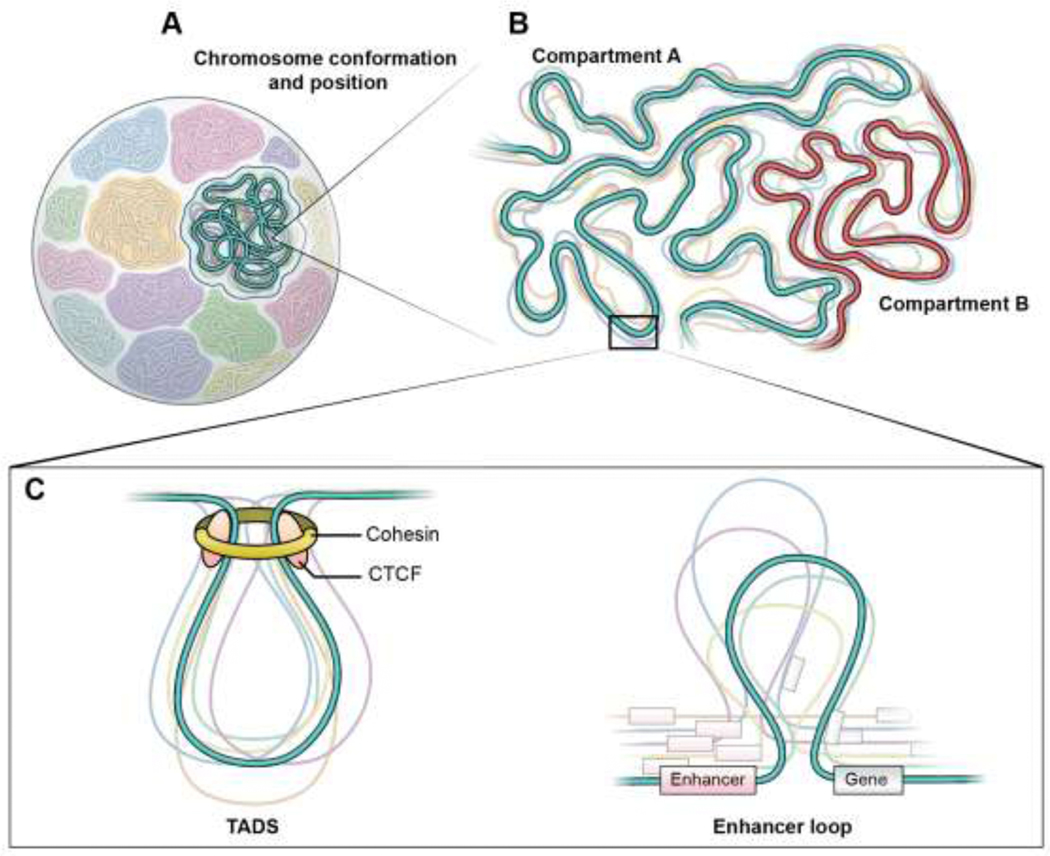
Figure 1: Extensive heterogeneity across all levels of genome organization:
(A) Within the eukaryotic nucleus the chromosomes occupy non-random territories but show variation their overall folding and their location between single cells. (B) Chromosomes are organized into mutually exclusive active and inactive compartments, referred as compartments A and B, respectively. Compartments across both the same and different chromosomes self-associate but vary in conformation and both the levels of compaction and segregation. (C) Compartments are further organized into loop domains that are driven by loop extrusion using the DNA motor protein, cohesin, and usually flanked by the insulator protein, CTCF, at their boundaries. These loops, referred as topological associated domains (TADs), show variation in loop size and boundary position between single cells. (D) TADs encompass smaller regulatory sub-loops that promote interactions of a gene with its distal enhancers. Such loops show high variability in size and anchor positions, resulting in variations in three-dimensional distances between enhancers and promoters amongst single cells.