FIGURE 1.
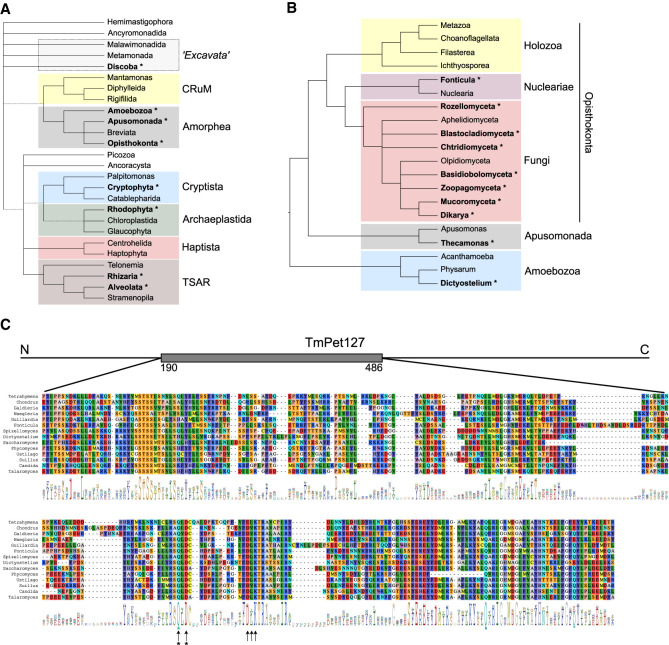
Pet127 is a conserved eukaryotic protein, lost in multiple independent lineages. (A) Presence of Pet127 in different eukaryotic supergroups. Names in bold with an asterisk indicate that at least one member of the group carries a Pet127 ortholog. Supergroups and putative tree topology are drawn following recent phylogenomic studies (Lax et al. 2018; Burki et al. 2020); dashed lines indicate uncertain monophyly of a group. Excavata are no longer considered a monophyletic supergroup. (B) Presence of Pet127 orthologs in Opisthokonta and related taxa. Tree topology follows the conclusions of phylogenomic analyses of Paps et al. (2013), fungal subkingdoms are according to Tedersoo et al. (2018). Ascomycete species, like S. cerevisiae, S. pombe, C. albicans, and T. marneffei, belong to Dikarya. (C) Alignment of Pet127p amino acid sequences from selected eukaryotic lineages (the full list of taxa and accession numbers can be found in Supplemental Table S1). Detailed alignment is shown for the most conserved core nuclease domain, its location in the sequence of Talaromyces marneffei Pet127 protein is marked on the schematic (gray box). Conservation is visualized as the sequence logo (Schneider and Stephens 1990) below the alignment, with the total height of the position indicating the degree of conservation measured as the information content on a scale of 0 to 4.3 bits. Arrows indicate the key amino acids in the conserved nuclease core (Kosinski et al. 2005; Steczkiewicz et al. 2012): E340, and the residues forming the variant of the PD-(D/E)XK motif (here QD-DLK): Q386, D388, D401, L402, K403. Residues mutated in the described experiments are marked with asterisks. Numbering follows that of the T. marneffei sequence.