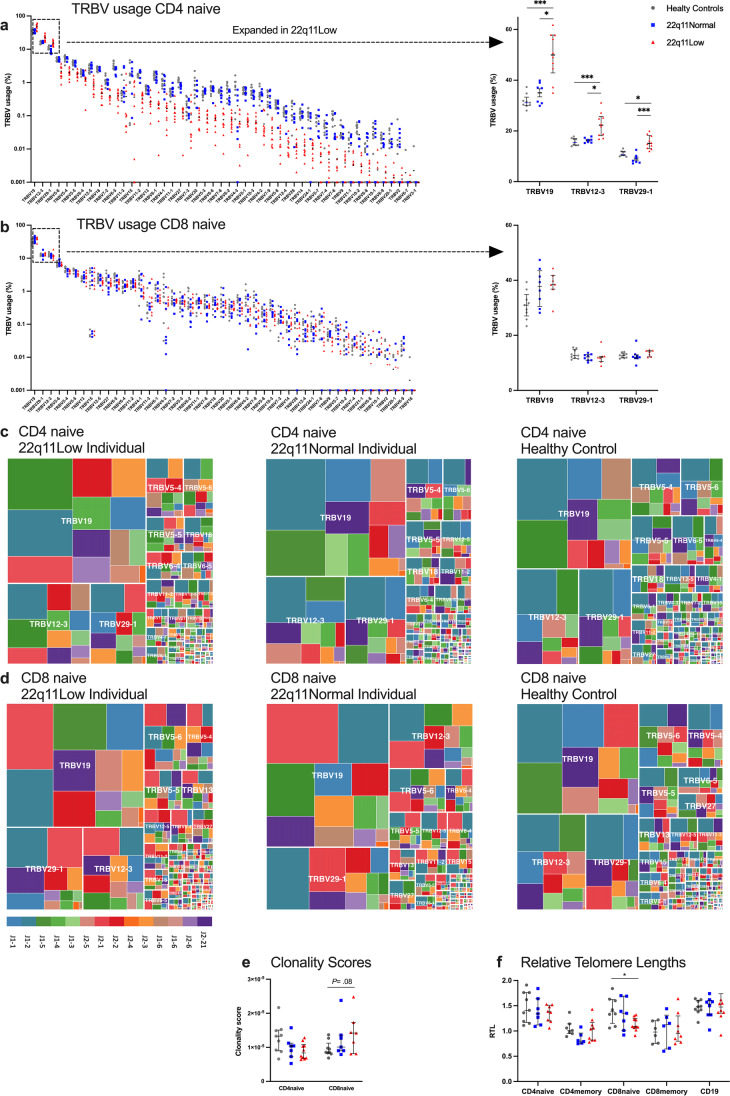
Fig. 2.
T-cell receptor repertoires and relative telomere lengths. TRBV gene usage in naïve T-helper lymphocytes (CD4 naïve) (a) and naïve cytotoxic T lymphocytes (CD8 naïve) (b). The TRBV genes are ordered from most-frequently used (left) to least-frequently used (right) in the 22q11Low group. Usage rates of the most frequent TRBV genes are presented in an inset to the far right. Only unique sequences (as defined by TRBV gene usage and amino acid sequence of the CDR3) were used for the T-cell receptor repertoire analysis. Tree plots representing the diversity and clonality of CD4 naïve cells (c) and CD8 naïve cells (d) in the 22q11Low individuals (left), the healthy control matched for age and gender (right), and the matched 22q11Normal individual (middle). The box size is proportional to the extent of V and J gene usage. Major TRBV genes are indicated on each tree plot, whereas TRBJ genes are identified by the color code at the bottom left. Clonality scores of the CD4 naïve and CD8 naïve cell subsets (e). Relative telomere lengths (RTL) in the naïve and memory subsets of CD4, naïve and memory CD8, and B lymphocytes (CD19) (f). Results are shown as individual values for the healthy controls (gray dots), 22q11Normal individuals (blue squares), and 22q11Low individuals (red triangles). Information on missing data is provided in Supplementary Table S3. Lines denote medians and whiskers indicate the interquartile ranges. P value summary indicated on each graph: *P ≤ .05, **P ≤ .01, ***P ≤ .001