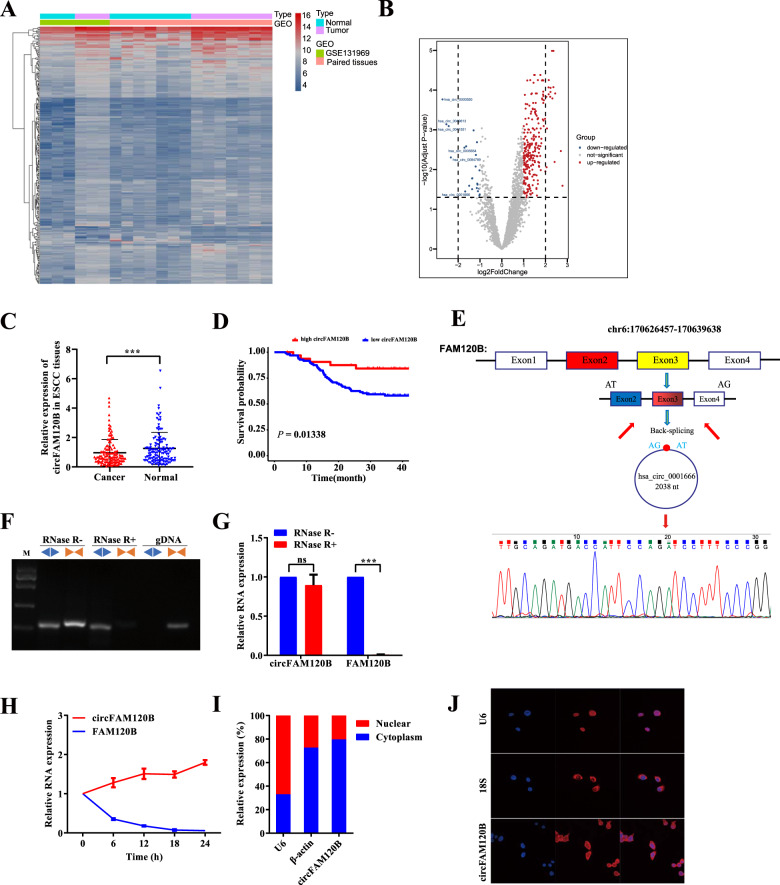
Fig. 1. Identification of circFAM120B as a circRNA in association with ESCC.
A, B The heatmap and volcano plot showed 276 differentially expressed circRNAs in ten paired ESCC tissues by Arraystar Human Circular RNA Microarray. The cutoff value was |log2(FC)| > 1 & Padjust < 0.05. C Detection of circFAM120B in an additional 130 paired ESCC tissues by qRT-PCR analysis. D Kaplan–Meier analysis of ESCC patients based on circFAM120B expression (n = 130). The upper quartile level of circFAM120B was defined as the cutoff value. A log-rank test determined statistical significance. E The back-splice junction site of circFAM120B was confirmed by Sanger sequencing. F, G circFAM120B in cDNA and genomic DNA were analyzed by PCR and 2% agarose gel electrophoresis using divergent primers or convergent primers, respectively. H Changes in the abundance of circFAM120B and FAM120B were analyzed after treatment with actinomycin D (5 μg/ml) at indicated time points. I Relative abundance of circFAM120B, β-actin, and U6 in the nuclear and cytoplasmic fractions of KYSE-150 cells was analyzed. J circFAM120B localization was analyzed by RNA FISH in KYSE-150 cells. ***P < 0.001.