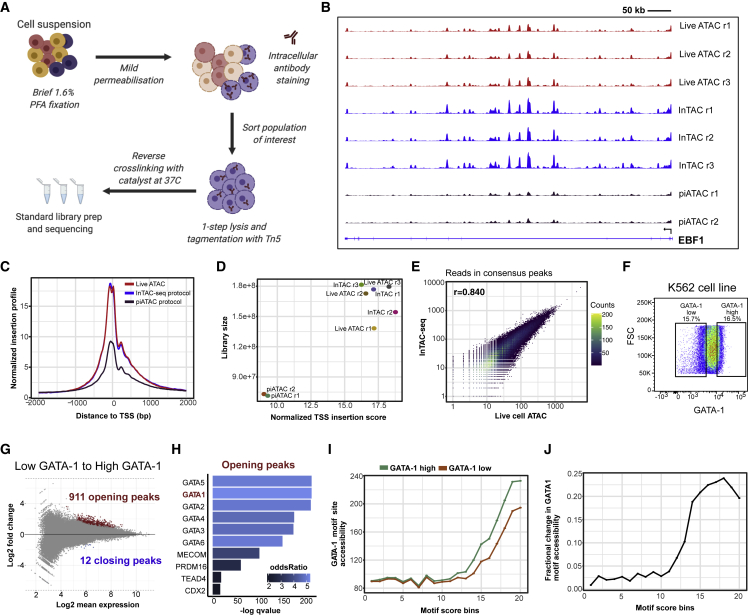
Figure 1.
InTAC-seq data from fixed cells is of comparable quality to ATAC-seq data from live cells and allows interrogation of chromatin accessibility associated with TF protein abundance
(A) Overview of InTAC-seq experimental protocol.
(B) Genome coverage of ATAC-seq data generated from live cells, fixed cells using InTAC-seq, or fixed cells using piATAC at the EBF1 locus in GM12878 cells.
(C) Normalized Tn5 insertion profiles centered at transcription start sites (TSSs) for the indicated ATAC-seq libraries.
(D) Scatterplot of estimated library size versus normalized TSS insertion score across all replicates of compared protocols.
(E) Scatterplot of reads in consensus peaks averaged across replicates between InTAC-seq and live ATAC samples, with calculated Spearman correlation coefficient as shown.
(F) FACS plot of forward scatter (linear scale) versus GATA-1 protein abundance (log10 scale) and the gating strategy to isolate the highest and lowest 15% of GATA-1-expressing K562 cells.
(G) MA plot of log2 fold change in accessibility between GATA-1-high and GATA-1-low K562 populations versus log2 mean number of reads at all consensus peaks. Peaks with significant changes in accessibility are highlighted in red or blue.
(H) Most significantly enriched TF motifs in differentially accessible peaks in GATA-1-high cells calculated using Fisher’s test.
(I) Average accessibility of GATA-1 motif sites across all consensus ATAC-seq peaks binned by GATA-1 motif score. Accessibility is defined here as the area under the curve of a plot of bias-corrected, normalized Tn5 insertions centered at GATA-1 motif sites (as in Figure S1G), integrated from −50 to +50 bp and excluding the TF footprint from −10 to +10 bp.
(J) Difference in GATA-1 motif accessibility between GATA-1 high and GATA-1 low samples normalized to the accessibility in the GATA-1 low population for each motif score bin.