Figure 2.
GATA-1-high BM progenitors show strong priming for erythroid lineage
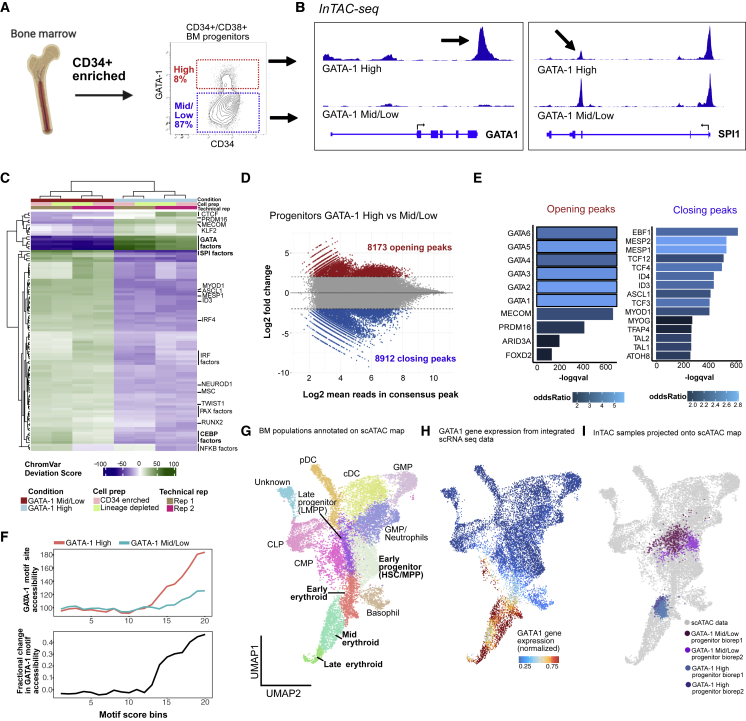
(A) BM aspirate is ficolled and enriched for CD34+ cells before gating for CD34+/CD38+ cells and selecting GATA-1-high population (top ∼8%) and remaining GATA-1-expressing cells (denoted as GATA-1 mid/low) (bottom ∼87%).
(B) InTAC-seq genome coverage plots at GATA1 and SPI1 loci for GATA-1-high and -mid/low BM progenitors.
(C) Heatmap of chromVAR deviation scores across GATA-1 high and mid/low BM progenitors for top 50 most variable motifs.
(D) MA plot of log2 fold change in accessibility between GATA-1 high and mid/low BM progenitors versus log2 mean number of reads in consensus peaks. Peaks with significant changes in accessibility are highlighted in red or blue.
(E) Most significantly enriched TF motifs in differentially accessible peaks between GATA-1-high and -mid/low BM progenitors calculated using Fisher’s test.
(F) (Top) Average accessibility of GATA-1 motif sites across all consensus ATAC-seq peaks binned by GATA-1 motif score. Accessibility is defined here as the area under the curve of a plot of bias-corrected, normalized Tn5 insertions centered at GATA-1 motif sites, integrated from −50 to +50 bp and excluding the TF footprint from −10 to +10 bp. (Bottom) Difference in GATA-1 motif accessibility between GATA-1 high and mid/low samples normalized to the accessibility in the GATA-1 mid/low population for each motif score bin.
(G) UMAP of previously published and annotated BM scATAC dataset with Seurat clusters manually annotated as key BM populations.
(H) Normalized GATA1 gene expression across BM progenitors in UMAP space (expression derived from scRNA-seq data integrated with scATAC-seq data).
(I) Bulk BM progenitor InTAC-seq data simulated as scATAC counts and projected onto scATAC UMAP space.