Figure 3.
High-dimensional, single-cell proteomic analysis of BM progenitors identifies TF and surface-marker trends in erythroid commitment
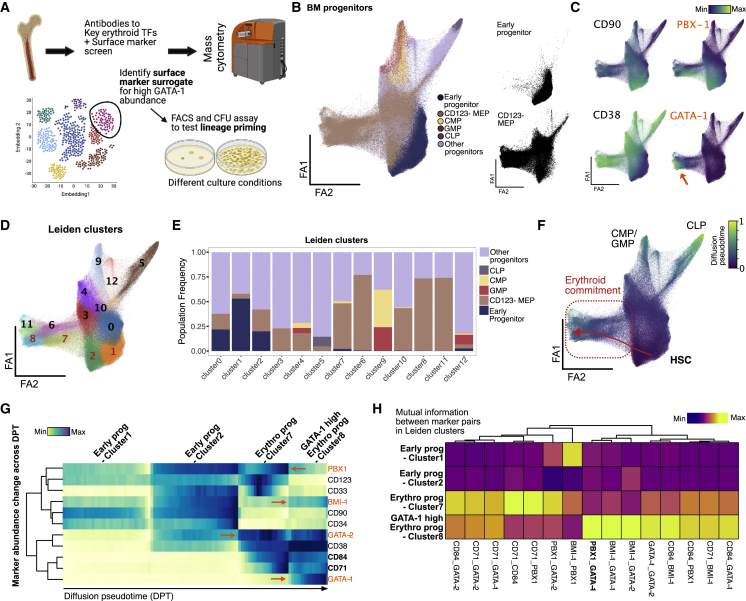
(A) BM aspirate is ficolled and enriched for CD34+ cells before staining with antibodies to surface marker panel and key TF to capture single-cell protein abundance using mass cytometry. High-dimensional data with over 1 million cells were used to delineate heterogeneity in BM progenitors and find surface-marker surrogates for GATA-1 TF abundance for further functional validation.
(B) Force-directed layout (ForceAtlas2) of density downsampled (to 250,000 cells) CD45+-gated BM progenitors colored by manually gated populations.
(C) Normalized marker expression of key surface and TF proteins (in orange) across force-directed layout. Orange arrow denotes GATA-1-high region in single-cell map.
(D) Leiden clusters of BM progenitors (resolution = 1) visualized on force-directed layout.
(E) Barplot of frequency of manually gated BM progenitor populations across 11 Leiden clusters.
(F) Normalized diffusion pseudotime calculation visualized on force-directed layout with trajectory from HSCs to erythroid-primed progenitors across Leiden clusters 3, 1, and 11 (in red).
(G) Row-normalized heatmap of median marker abundance at 100 bins across diffusion pseudotime-aligned trajectory. Orange font and arrows indicate TF protein trends, and bold font with black arrows indicate key surface marker trends in trajectory.
(H) Column-normalized heatmap of mutual information scores calculated on cells in Leiden clusters 2, 7, and 8 and normalized across key TF and surface-marker pairs.