Figure 4.
Estimation of KD values by RBNS is robust and reproducible
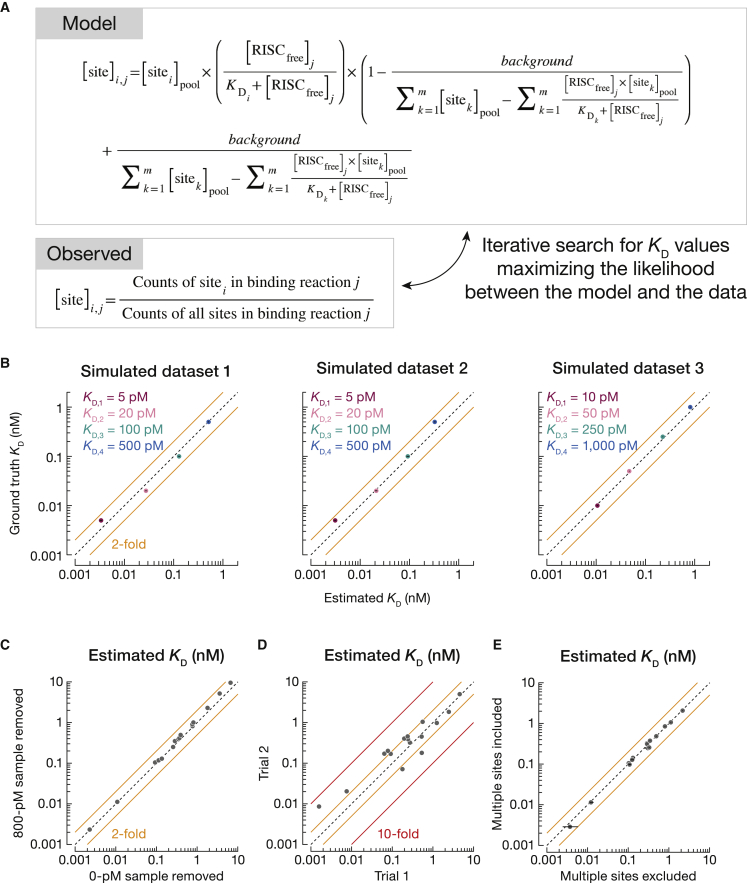
(A) The mathematical equation derived from the biochemical model at equilibrium describes predicted concentration of binding site i in sequencing data, given KD values of all binding sites, miRISC concentration, and background. These parameters are fit simultaneously to maximize the likelihood of observing predicted sequencing counts in experimental data.
(B) Testing KD estimation with simulated data. RBNS data were modeled by simulating miRISC binding to RNA pool containing four binding sites and no-site molecules. Stock concentration of miRISC was equal to 2.1 nM (dataset 1) or 8.1 nM (datasets 2 and 3). Background was set to 0.1 nM and KDnosite was set to 5 nM. Error bars indicate 95% CI on the median.
(C) Comparison of sub-datasets when the highest miRISC concentration and no-RISC binding reaction were removed (Pearson’s r = 0.979). Binding sites identified in Figure 3 were used to compute KD.
(D) Correspondence between fitted KD values of enriched binding sites estimated from two independent binding experiments (Pearson’s r = 0.974). The solid orange and red lines indicate 2- and 10-fold differences, respectively. Dashed diagonal lines show y = x.
(E) Comparison of fitted KD values when multi-site reads were fractionally assigned to corresponding site types or excluded from the analysis.