Figure 1.
Development of HT-5Pseq
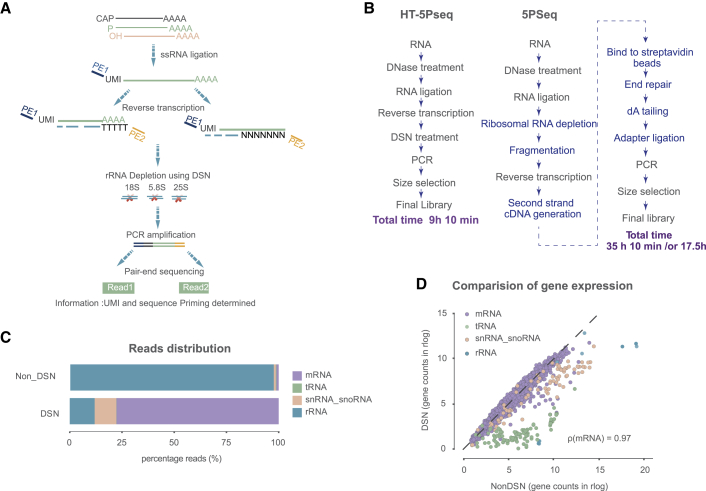
(A) Outline of HT-5Pseq method . A specific RNA oligo (PE1) is ligated to 5′P RNA molecules. RNA is reverse transcribed by using a mix of sequencing oligos (PE2) containing oligo(dT) and random hexamers. The cDNA originating from rRNA is depleted using DNA oligos and double-strand specific nuclease (DSN). cDNA is PCR amplified and sequenced.
(B) Flowchart of HT-5Pseq and 5Pseq for each step. Different and eliminated steps in 5Pseq are marked in blue.
(C) Improvement of mRNA mappability in S. cerevisiae HT-5Pseq after rRNA depletion. NonDSN refers to control libraries omitting rRNA depletion probes.
(D) Differential gene-specific 5′P read coverage.