Figure 2.
HT-5Pseq reveals ribosome dynamics at codon resolution
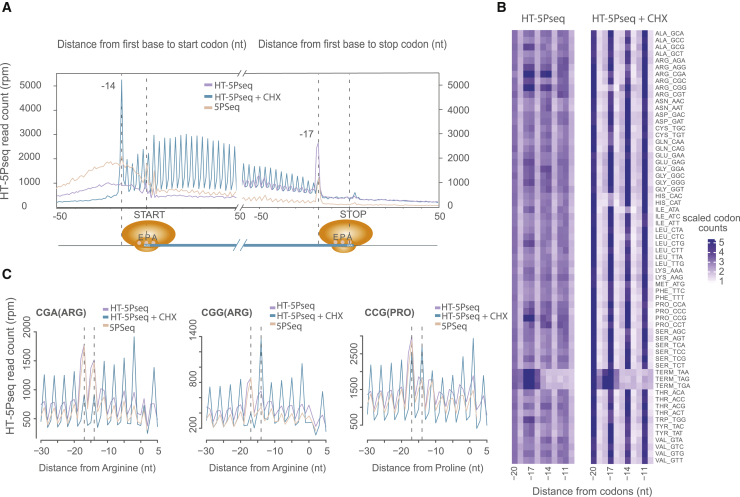
(A) Metagene analysis for 5′P read coverage relative to open reading frame (ORF) start and stop codon. Cells grown in rich medium (YPD) by HT-5Pseq are shown in purple, cells treated with cycloheximide (CHX) for 5 min in blue, and 5Pseq cells in pink.
(B) Heatmap for codon-specific 5′P coverage. Positions −17, −14, and −11 nt represent ribosome protection at the A, P, and E site, respectively, with or without CHX treatment. For each codon, reads were normalized using the total 5′P reads between −30 and 5 nt.
(C) 5′P reads coverage for rare arginine (CGA, CGG) and proline codons (CCG). Dotted lines at −17 and −14 correspond to the expected 5′ end of protected ribosome located at the A site or P site, respectively.