Figure 3.
Regulation of termination associated ribosome pauses
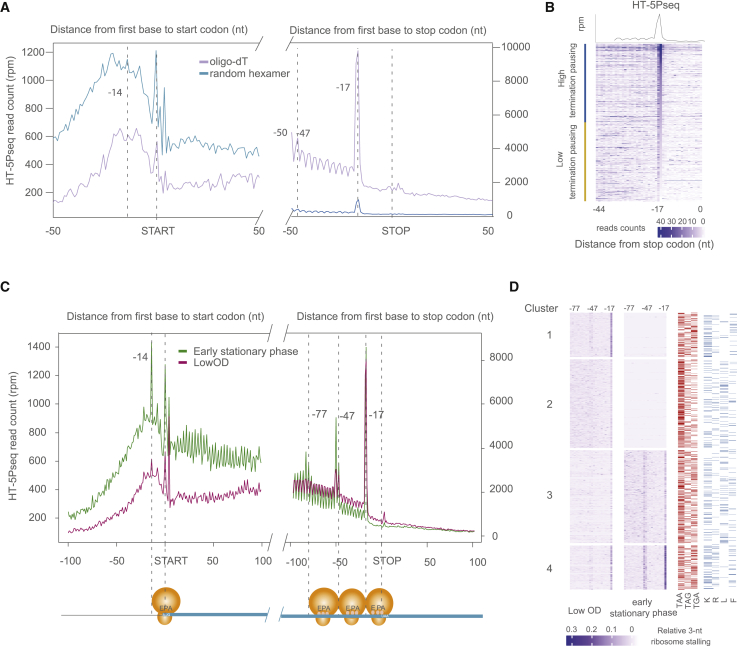
(A) Metagene analysis for 5′P read coverage relative to ORF start and stop codon for HT-5Pseq reads primed by oligo(dT) (purple) or random hexamer (blue).
(B) Gene-specific 5′P read coverage relative to ORF stop codon. Genes are sorted by the relative ribosome protection at the stop codon (−17) with respect to the upstream region (−44 to −17 nt). Only genes with at least 10 rpm (reads per million) in this region were considered. The top 50% genes were defined as the high termination pausing genes and the bottom half were defined as low termination pausing genes.
(C) Metagene analysis for 5′P read coverage relative to ORF start and stop codon. Cells were grown under different growth conditions. Exponentially growing cells at a lower cell density (OD600 ~ 0.3) are shown in purple and early stationary phase in green.
(D) Relative 3-nt periodicity for the last 12 codons clustered using k-means. Each point corresponds to the ratio at each codon (ratio between peaks and valleys), not individual 5′P coverage as in (B). −17 nt indicates a ribosome paused at the stop codon and −50 nt corresponds to disome protection. The stop codon usage is represented in red and amino acids usage before stop codons (K, R, L, F) in blue.