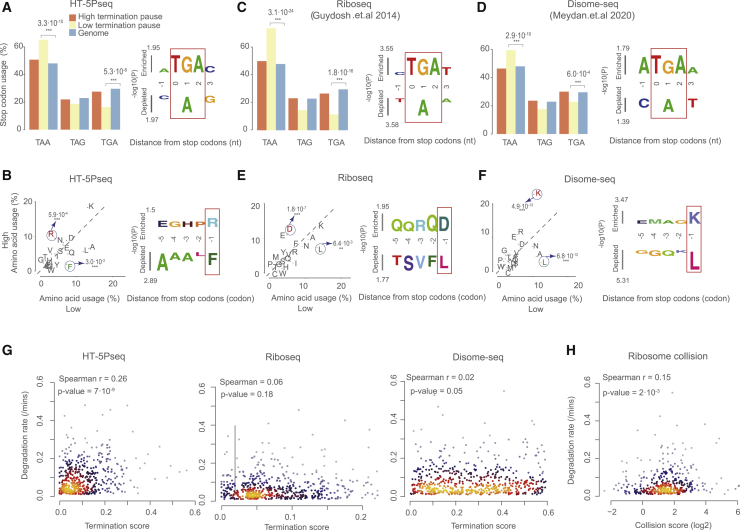
Figure 4.
Stop codon context at termination pausing associated with mRNA decay
(A) Frequency of stop codons usage in genes (left) and significance of enrichment for particular nucleotides at each position relative to stop codon (right) using kpLogo (Wu and Bartel, 2017) by comparing high and low termination pausing groups in HT-5Pseq. Significant deviation from genome average was estimated using a hypergeometric test; only significant p values are shown.
(B) Frequency of last amino acid usage in genes with high termination pausing (y axis) versus low termination pausing (x axis) (left) and significance of enrichment for particular amino acids at each position relative to stop codon (right) as in (A).
(C and D) As (A) but for (C) ribosome profiling data and (D) disome profiling data from Guydosh and Green (2014) and Meydan and Guydosh (2020).
(E and F) As (B) but for (E) ribosome profiling data and (F) disome profiling data.
(G) Spearman correlation between RNA degradation (calculated by total mRNA half-life from Presnyak et al., 2015) with termination pause in HT-5Pseq (left), ribosome profiling (middle), and disome profiling (right) data.
(H) Spearman correlation between RNA degradation with collision scores. Collision scores were calculated from the disome profiling dataset by dividing disome peaks and monosome peaks of terminations stalling at stop codons.