Figure 2.
scMM analysis on the PBMC CITE-seq dataset
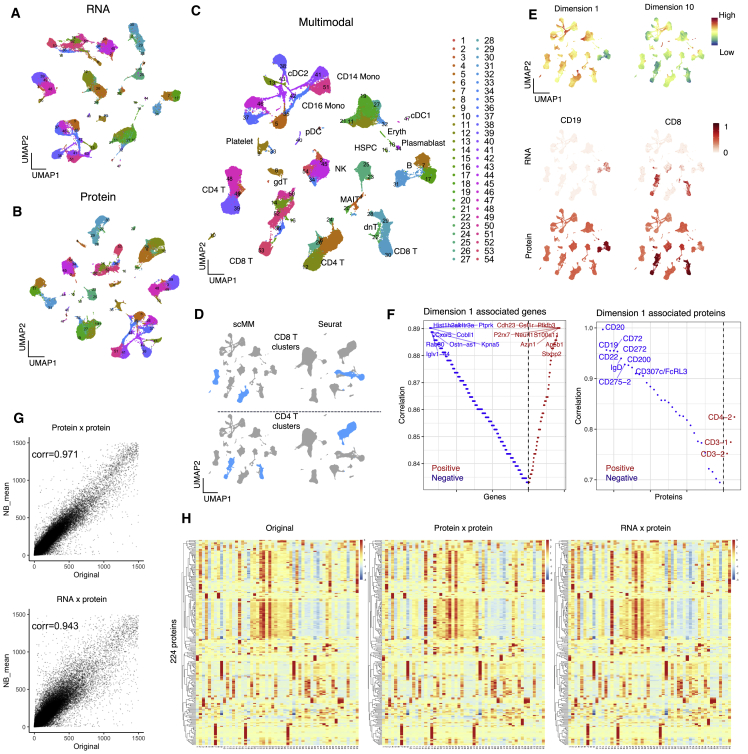
(A–C) UMAP visualization of unimodal latent variables for transcriptome, surface protein, and multimodal latent variables, respectively. Each dot represents a single cell and is color coded according to the clustering performed on multimodal latent variables.
(D) CD8 and CD4 T cells annotated in scMM and Seurat analysis are color coded in the UMAP visualization.
(E) Top: multimodal UMAP visualization colored according to the latent dimension values. Shown in the middle and on the bottom is a UMAP visualization colored according to the transcriptome and protein counts for cell type markers, respectively.
(F) Genes (left) and surface proteins (right) associated with latent dimension 1. Each feature was aligned on the basis of the Spearman correlation coefficient. The y axis represents the absolute correlation coefficient, and red and blue represent positive and negative correlations, respectively.
(G) NB mean parameters reconstructed from surface protein or transcriptome counts were plotted against original surface protein counts for each cell. The mean parameters were plotted for all available proteins. Pearson correlation coefficients are shown in the plots.
(H) Heatmap constructed from the original (left), unimodal generation (middle), and crossmodal generations. Rows and columns represent the measured 224 surface proteins and 54 clusters discovered by PhenoGraph, respectively.