Figure 3.
Prediction of surface protein measurements for the BMNC dataset
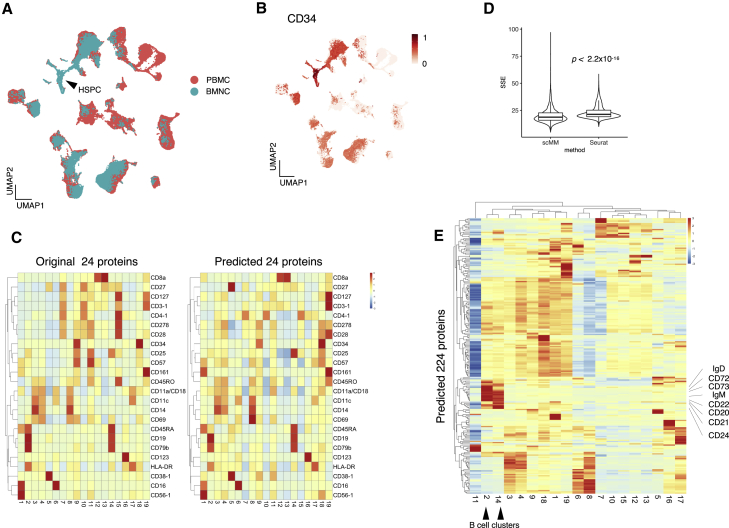
(A) Joint UMAP embedding of transcriptome latent variables inferred from PBMC training data and the BMNC dataset. The black arrowhead indicates the HSPC population.
(B) UMAP embedding of the BMNC dataset was colored according to the protein expression levels of CD34.
(C) Heatmaps constructed from original (left) and predicted (right) surface protein counts. Rows and columns represent 24 shared surface protein markers and clusters discovered by PhenoGraph, respectively.
(D) Benchmarking on surface protein prediction performance of scMM against Seurat. Centered log-transformed data were used to calculate the sum of squared error (SSE) per cell and plotted for each prediction result. Statistical analysis was performed with the two-sided Wilcoxon signed-rank test.
(E) Heatmap constructed with the predicted 224 surface protein markers. Rows and columns represent surface protein markers and clusters, respectively. Black arrowheads denote B cell clusters, and their markers are indicated.