Figure 4.
scMM analysis on mouse skin SHARE-seq dataset
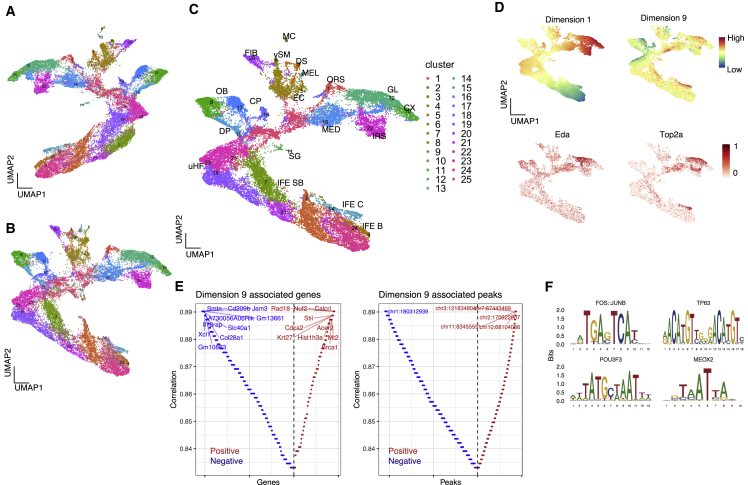
(A–C) UMAP visualization of unimodal latent variables for transcriptome, chromatin accessibility, and multimodal latent variables. Each dot represents a single cell and is color coded according to the clustering performed on multimodal latent variables.
(D) Top: multimodal UMAP visualization color coded on the basis of latent dimension values. Shown in the middle and on the bottom is a UMAP visualization colored according to transcriptome counts for cell type markers.
(E) Genes (left) and peaks (right) associated with latent dimension 9. Each feature was aligned on the basis of Spearman correlation coefficient. The y axis represents the absolute correlation coefficient, where red and blue colors represent positive and negative correlations, respectively.
(F) Motif plot for representative motifs significantly enriched in peaks positively associated with latent dimension 9.