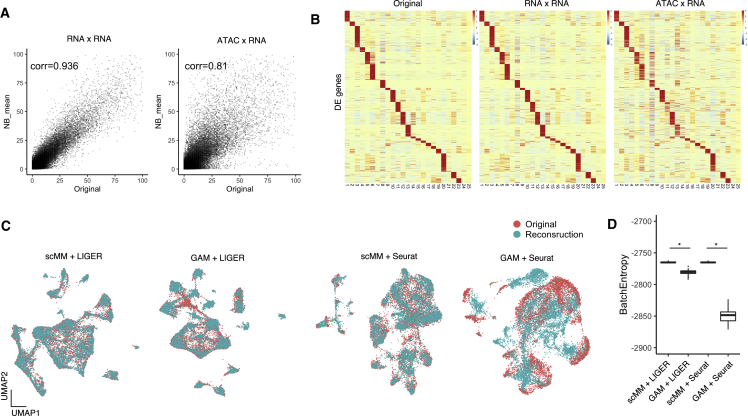
Figure 5.
Crossmodal generation from chromatin accessibility to transcriptome leads to better data integration
(A) NB mean parameters reconstructed from transcriptome or chromatin accessibility counts are plotted against original transcriptome counts for each cell. Pearson correlation coefficients are shown in the plots.
(B) Heatmaps constructed from original, unimodal generation, and crossmodal generation transcriptome data. Rows and columns represent deferentially expressed (DE) genes and clusters discovered by PhenoGraph, respectively.
(C) Joint visualization of original and predicted single-cell transcriptome data integrated by LIGER and Seurat. For the prediction, either scMM or GAM was used.
(D) Boxplot showing entropy of batch mixing for each integration. Statistical test performed with two-sided Wilcoxon rank-sum test. ∗p < 2.2 × 10−22.