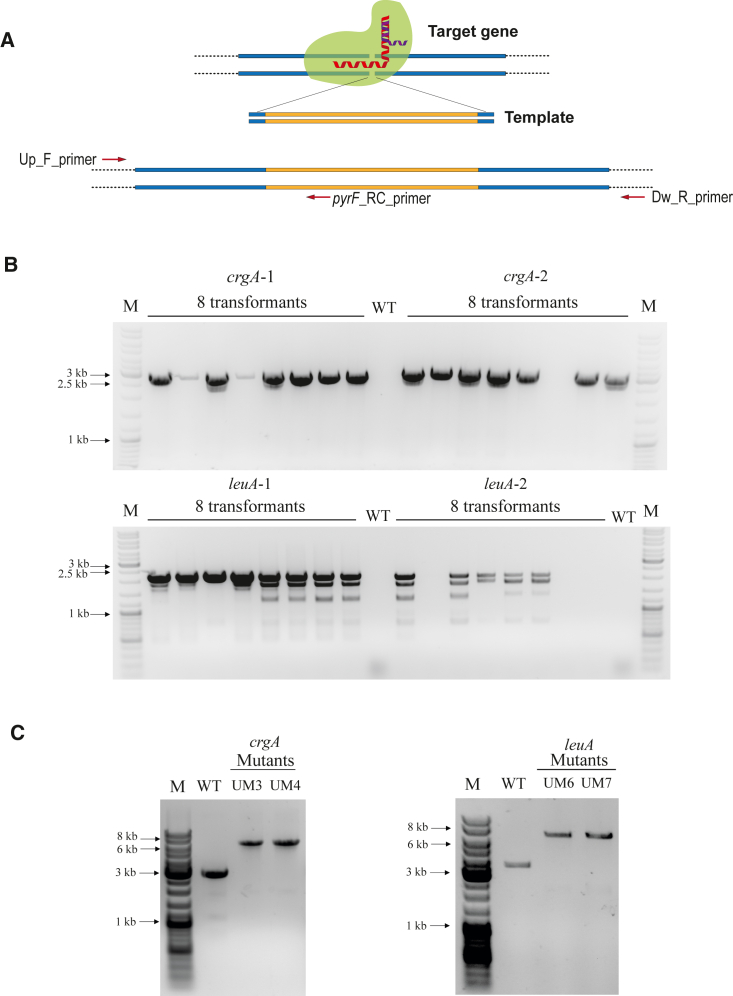
Figure 3.
Disruption of crgA and leuA genes
(A) Strategy followed to disrupt target genes with the combination of gRNA-Cas9 complex and a designed DNA template. Positions of the primers used to check gene disruption and homokaryosis are also shown.
(B) Gene disruption validation by PCR using the Up_F_primer and the pyrF_RC_primer for eight randomly selected transformants obtained with crgA_gRNA1 + crgATemplate1 (expected fragment of 2.8 kb), crgA_gRNA2 + crgATemplate2 (expected fragment of 2.9 kb), leuA_gRNA1 + leuATemplate1 (expected fragment of 2.1 kb), and leuA_gRNA2 + leuATemplate2 (expected fragment of 2.2 kb). DNA from the R. microsporus wild-type strain ATCC 11559 was used as a negative control (WT).
(C) PCR fragments smaller than the ones expected in some lanes of the leuA transformants corresponded to nonspecific PCR products. (C) PCR amplification of the crgA and leuA locus of the indicated transformants using the Up_F_primer and the Dw_R_primer. Expected PCR DNA fragments from WT and disrupted crgA locus were 3.1 kb and 6.6 kb in length, respectively. For leuA, expected PCR products from WT and disrupted locus were 3.6 kb and 7.1 kb in length, respectively. Black arrows point to the indicated bands of the marker.