Figure 1.
Bioinformatic identification of genomic safe harbor sites
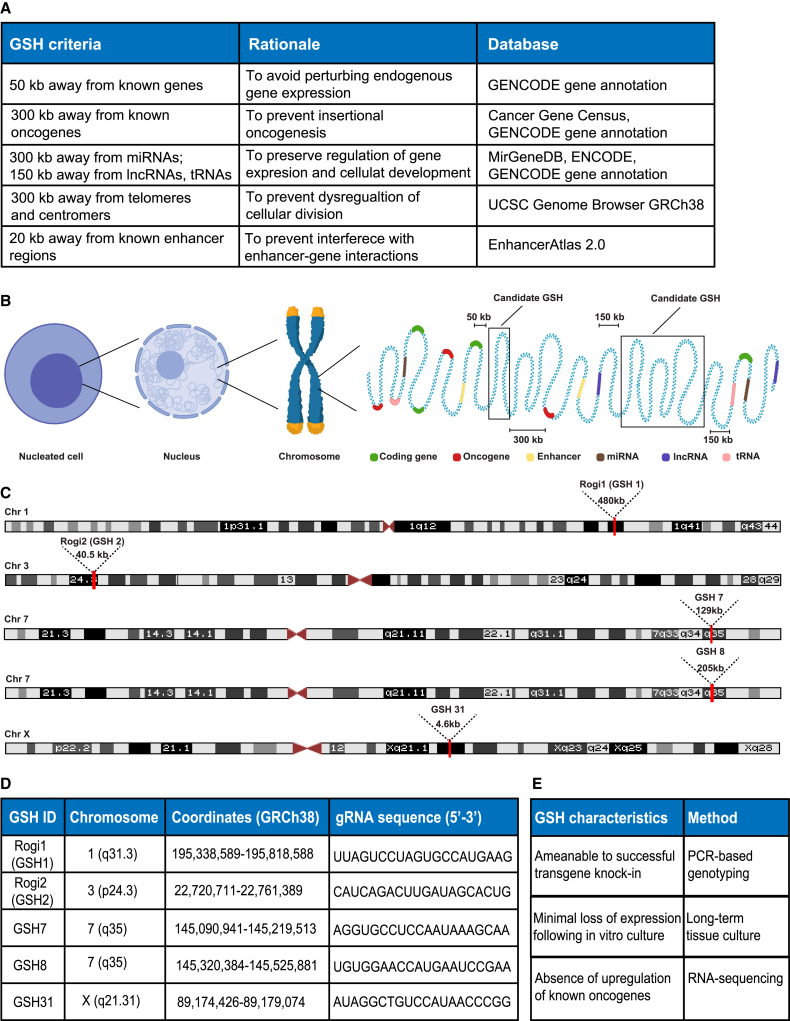
(A) Table shows GSH criteria, rationale, and databases used to computationally predict GSH sites in the human genome.
(B) Schematic representation of candidate GSH sites, showing linear distances from different coding and regulatory elements in the genome according to the established and newly introduced criteria.
(C) Chromosomal locations and lengths of 5 candidate GSH sites, which were subsequently experimentally tested.
(D) Chromosomal coordinates of 5 candidate GSH sites and the gRNA sequences used for subsequent CRISPR/Cas9 genome editing. See also Table S1 for the list of all of the computationally predicted sites.
(E) Characterization of experimentally tested GSH sites includes genotyping of the targeted locus to verify transgene insertion, long-term culture of GSH integrated cells to observe the durability of transgene expression, and bulk and single-cell RNA sequencing (RNA-seq) to validate safety of integration by absence of upregulation of genes associated with oncogenic cellular proliferation.