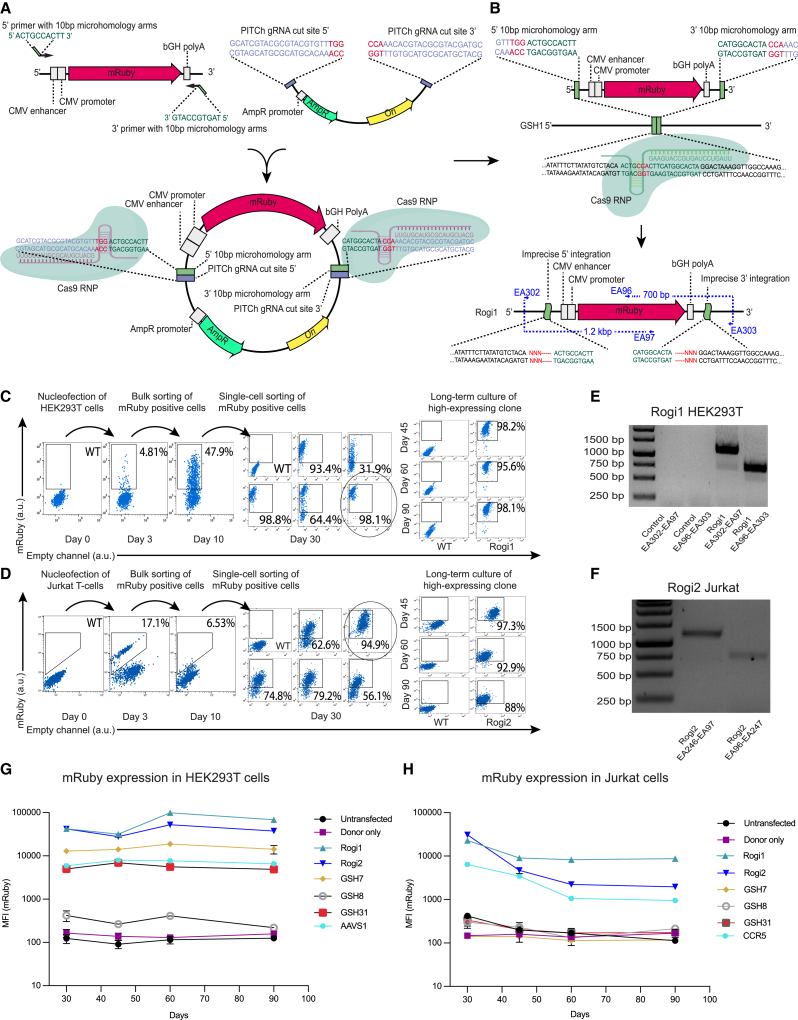
Figure 2.
Experimental validation of candidate GSH sites by targeted genome insertions in HEK293T and Jurkat cells
(A) Generation of PITCh plasmid by cloning an mRuby-bearing insert with microhomologies against specific GSH into a backbone possessing PITCh gRNA target sites, required for the Cas9-based liberation of the insert.
(B) mRuby insert is integrated into a desired site by the MMEJ pathway following a Cas9-induced double-stranded break.
(C and D) Flow cytometry demonstrating the isolation of clonal populations expressing the mRuby transgene from Rogi1 locus in HEK293T cells and Rogi2 locus in Jurkat cells using pooled and single-cell flow cytometry-mediated sorting. The highest expressing Rogi1-HEK293T clone and the Rogi2-Jurkat clone were expanded in cell culture, and flow cytometry measurements at days 45, 60, and 90 demonstrated stable levels of transgene expression.
(E) Genotyping of the Rogi1 site in HEK293T cells using primers spanning the junction between integration site and the transgene. Lane names refer to the primers used from (B). Control and Rogi1 EA302-EA97 lanes correspond to the 5′ integration junction in untransfected HEK293T cells and cells that were transfected using the PITCh CRISPR/Cas9 method, respectively. Control and Rogi1 EA97-EA303 lanes correspond to the 3′ integration junction in untransfected HEK293T cells and cells that were transfected using the PITCh CRISPR/Cas9 method, respectively.
(F) Genotyping of the Rogi2 site in Jurkat cells using primers spanning the junction between integration site and the transgene. The Rogi2 EA246-EA97 lane corresponds to the 5′ integration junction, while EA96-EA247 corresponds to the 3′ integration junction in HEK293T cells that were transfected using the PITCh CRISPR/Cas9 method.
(G) Continuous assessment of mRuby expression levels following the integration into each of the tested GSH sites as well as into AAVS1 control in HEK293T cells. Data are represented as means ± SEMs of the highest expressing clonal population of each tested site; N = 2.
(H) Continuous assessment of mRuby expression levels following the integration into each of the tested GSH sites as well as into CCR5 control in Jurkat cells. Data are represented as means ± SEMs of the highest expressing clonal population of each tested site; N = 2.