Figure 2.
Characteristics of OP-Strand-seq libraries
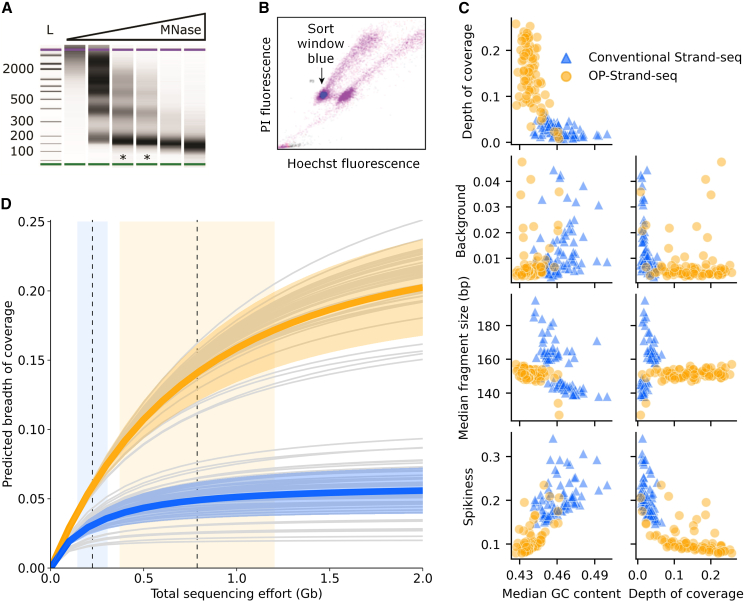
(A) DNA fragment size of nuclei digested with MNase (0–5,000 gel units/mL). The sizes of ladder bands (L) are given in base pairs. The lanes marked (∗) are correctly digested.
(B) A mixture of nuclei from human cells cultured with or without BrdU and stained with propidium iodide (PI) and Hoechst dye. Nuclei with BrdU show quenched Hoechst fluorescence.
(C) Properties of 78 OP-Strand-seq libraries and 78 conventional Strand-seq libraries. We used the outer coordinates of fragments from unique, properly paired reads to calculate the GC content, fragment length, and coverage. We used breakpointR (Porubsky et al., 2020b) to calculate the background, and we calculated spikiness (see results and discussion) as described in Bakker et al. (2016). The two clusters of conventional Strand-seq libraries in the fragment size plots are from different batches.
(D) Complexity curves for individual libraries (gray lines) produced with OP-Strand-seq or conventional Strand-seq, and their mean and SD (colored lines and ribbons). The dashed vertical lines and bands represent the means and SDs of the sequencing effort used (OP-Strand-seq on right, conventional on left). Breadth of coverage is the fraction of the haploid reference genome covered by at least one read fragment. Complexity estimates were made using Preseq (Daley and Smith, 2014).