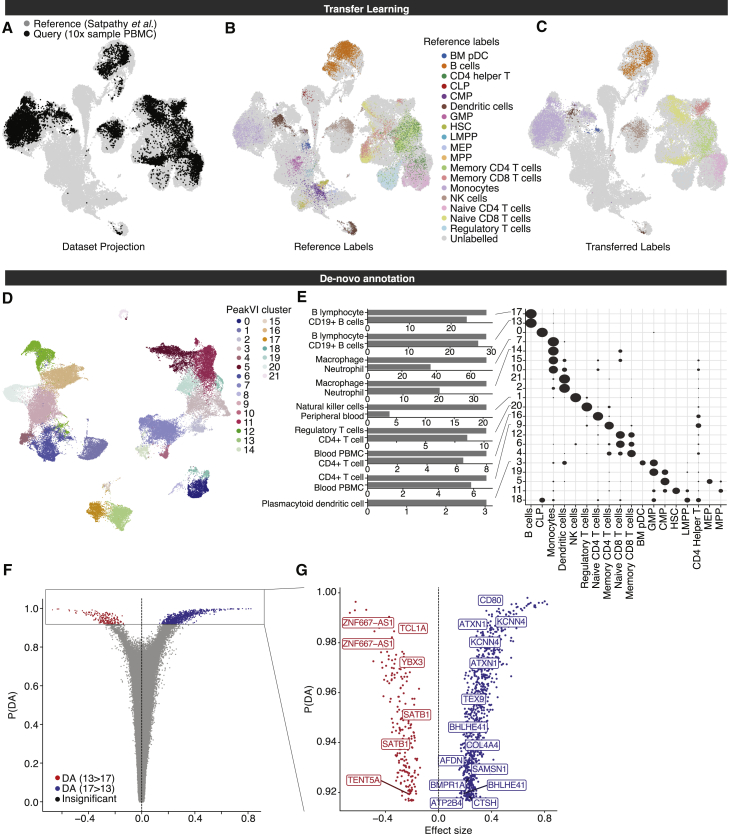
Figure 4.
PeakVI unlocks multiple paths for annotation and identification
(A–C) PeakVI supports transfer learning. (A) Mapping of query data (sample PBMC data from 10× Genomics) onto reference data (from Satpathy et al., 2019). PeakVI mixes the query data with the reference despite the data being generated by a different protocol and processed by a different pipeline. (B) The reference data, colored by FACS-based cell-type-specific labels. (C) The query data, colored by the transferred cell-type-specific labels.
(D–F) De novo annotation using PeakVI’s differential accessibility analysis. (D) Hematopoiesis data colored by clusters. (E) Regions that are preferentially accessible in each cluster were analyzed for enriched cell-type signatures from ARCHS (Lachmann et al., 2018) signatures, using enrichr (Chen et al., 2013; Kuleshov et al., 2016). Heatmap shows distribution of cell-type-specific labels for each cluster, normalized by row. (F) Volcano plot for a differential accessibility analysis between the two B cell clusters (clusters 13 and 17).
(G) Volcano plot for only significant regions, labeled by associated genes that are implicated in naive B cells (red) and memory B cells (blue).
See also Figures S4B and S4C.