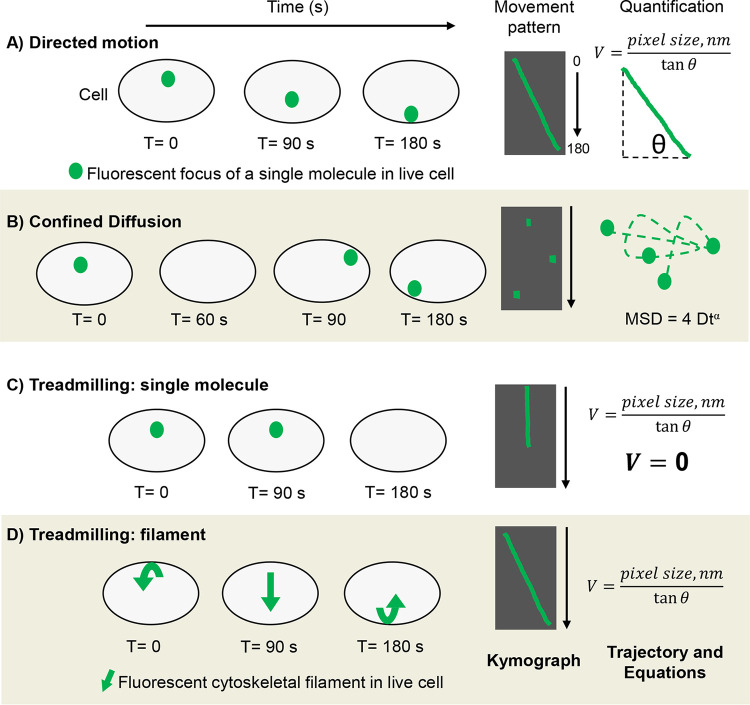
FIG 2.
Methods of representing and quantifying molecular movement patterns. Illustration of the cell body (gray with black outline) and single fluorescent foci (green dots) imaged using high-resolution microscopy. (A) Directed motion over 180 s and resulting kymograph showing focus trajectory (green) against cell body background (black for contrast). Kymographs can be generated using ImageJ and represent motion in 2D by stacking video pixels chronologically. For quantification, the velocity of the molecule displaying directed motion can be delineated by using the equation shown (nm = nanometer). (B) Diffusive motion captured using “fast” acquisition rate and resulting kymograph. Observe that kymographs are insufficient in representing diffusion because diffusive molecules do not form linear trajectories and often leave the plane of vision. Loss of visualization is illustrated as a blank cell in T = 60 and black gaps in kymographs. For quantification, the mean squared displacement (MSD) can be determined and plotted from raw image files using ImageJ. Analyzing MSDs reveals diffusion coefficients (diffusive molecules) as well as velocities (directed motion). (C) Single molecule within a treadmilling filament. Here, the molecule’s zero change in distance over time results in a straight-line trajectory. As expected, calculating the velocity using tanΦ of zero, results in V = zero, thereby adding mathematical confidence to the observed stagnation. (D) Directional movement of a treadmilling polymeric cytoskeletal filament (green arrow). A filament’s treadmilling rate is easily extrapolated by solving for V (velocity) or plotting the MSD.