FIG 2.
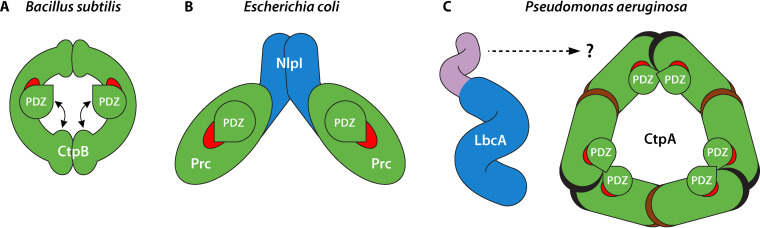
Contrasting arrangements of bacterial CTPs. (A) B. subtilis CtpB forms a ring-like dimer via N-terminal to N-terminal and C-terminal to C-terminal interactions. The positioning of the PDZ domain controls access to a narrow protease tunnel (red). CtpB switches between inactive and active forms, defined by whether or not the PDZ domain blocks the protease tunnel. (B) E. coli Prc is a monomer that forms a self-compartmentalized bowl-like structure, with part of the protease domain comprising a vault (red), and a lid-like PDZ domain. Upon interaction with a substrate, the PDZ domain shifts position causing a productive repositioning of the active site residues (not shown). The adaptor protein NlpI is a dimer, with each subunit interacting with one Prc monomer to form the tetrameric arrangement shown. NlpI is required for Prc to degrade MepS, but not for the degradation of any other Prc substrates in vivo. (C) P. aeruginosa CtpA forms a ring-like hexamer, arranged as a trimer of dimers mediated by N-terminal to N-terminal (black) and C-terminal to C-terminal (brown) interactions. The LbcA adaptor protein is a monomer with 11 TPR motifs forming a spiral structure (blue), and a separate N-terminal helical domain (purple) that is required for interaction with CtpA. LbcA is required for CtpA to degrade all five of its known substrates. The CtpA hexamer is inactive in the absence of LbcA because the active site residues (red) are misaligned (not shown). Studies to understand exactly how LbcA interacts with and activates CtpA are in progress.