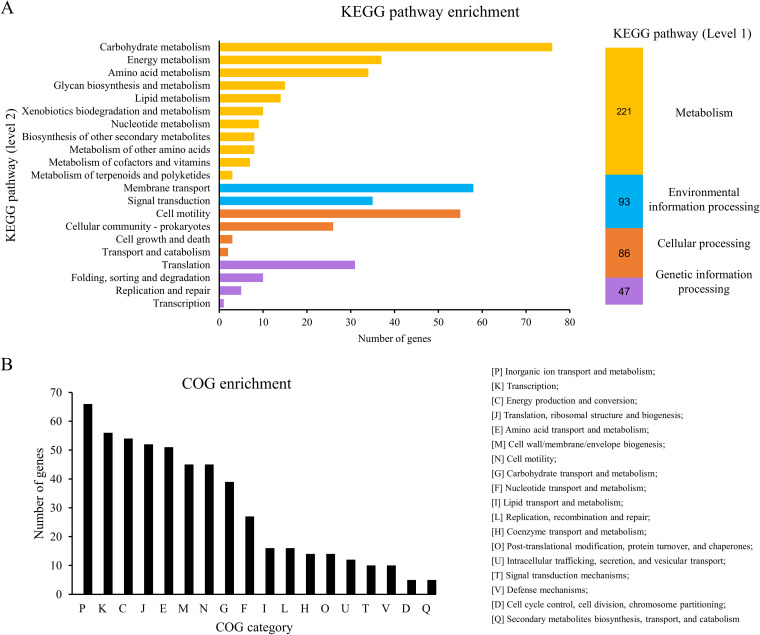
FIG 1.
Differentially expressed genes (Δtat versus WT). (A) KEGG pathway enrichment of the differentially expressed genes. The differentially expressed genes were analyzed by using the BlastKOALA tool (https://www.kegg.jp/blastkoala/) to assign K numbers which were then used for pathway enrichment by using KEGG Mapper (https://www.genome.jp/kegg/mapper/search.html). The numbers inside the vertical bars are the total numbers of genes belonging to each level 2 KEGG pathway. (B) COG enrichment of the differentially expressed genes. COG enrichment was performed with the EggNOG v5.0 database (http://eggnog5.embl.de/#/app/home).