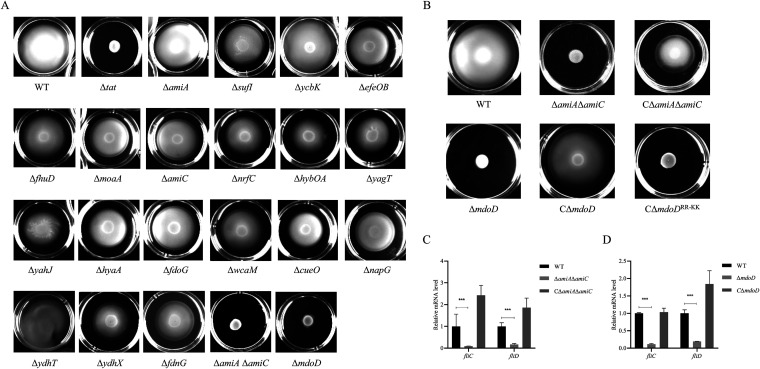
FIG 3.
Identification of Tat substrate proteins responsible for loss of motility. (A) For the swimming assay with the Tat substrate-related mutants, 5 μL of cells of each indicated strain grown to the mid-log phase was spotted onto the swimming plate (tryptone, 10 g/L; yeast, 5 g/L; NaCl, 10 g/L; agar, 0.2%) and was dried at room temperature for 5 min followed by incubation at 37°C for 7 h. The assay was performed in triplicate, and the data shown are representative. (B) Swimming assay with ΔamiAΔamiC, ΔmdoD, and the strains in trans expressing the corresponding proteins. The CΔmdoD and CΔmdoDRR-KK strains are the ΔmdoD strain encompassing a plasmid encoding the wild type and the mutated MdoD in which the twin-arginine in the signal peptide is replaced with a twin-lysine. CΔamiAΔamiC is the ΔamiAΔamiC strain in trans expressing AmiA and AmiC. (C) and (D) RT-qPCR analysis. Total RNA was extracted from cells of each indicated strain grown to the mid-log phase. Genomic DNA was removed, and cDNA was synthesized and was used as the template for qPCR with a QuantStudio 6 Flex fluorescence quantitative PCR instrument. The expression level of the tested gene was calculated using the 2−ΔΔCT method and normalized to the housekeeping gene gapA. Data are displayed as the geometric mean ± SEM. Statistical significance between the two groups was analyzed using Student’s t test. ***, P < 0.001.