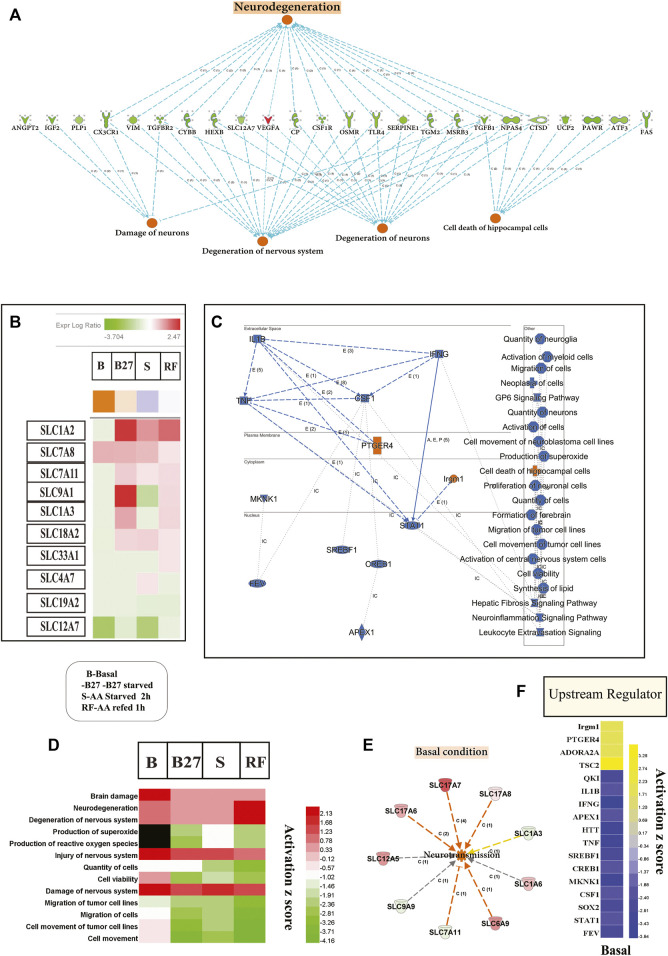
FIGURE 5.
The absence of SLC3A10 leads to transcriptome changes of genes in KO PCCs involved in modifying cellular, molecular functions and indicating neurological disease. Complete transcriptomic data and changed gene list subjected to IPA software and focused analysis performed on neurological function and disease. (A) Representation of genes from a basal level of transcriptomic data indicated a rise in neurodegeneration of KO PCCs (B). Heat-map representation of solute carrier transporter involved in neurodegeneration and affected expression under B, B27, S, and RF levels. (C) Repre-sentation of overall summary changes of molecules of Transcription regulator (TR), cytokine-Kinase-K- Enzyme- E, G-protein coupled receptor-GPCR, at a subcellular level, cellular, molecular functions, and cellular pathways in the basal level of KO PCCs. (D) Heat-map represents comparison analysis using IPA focused on disease, and cellular function at B, B27, S, and RF conditions accordingly activated Z scores. (E) At altered basal expression, SLC enhances neurotransmis-sion function in KO PCCs. (F) The heat map of top upstream molecules accordingly activated Z-scores, resulting in in-hibition and upregulation prediction at the basal level. Transcription regulator (TR), cytokine-Kinase-K- Enzyme- E, G-protein coupled receptor-GPCR. Upstream regulator analysis was performed using IPA.