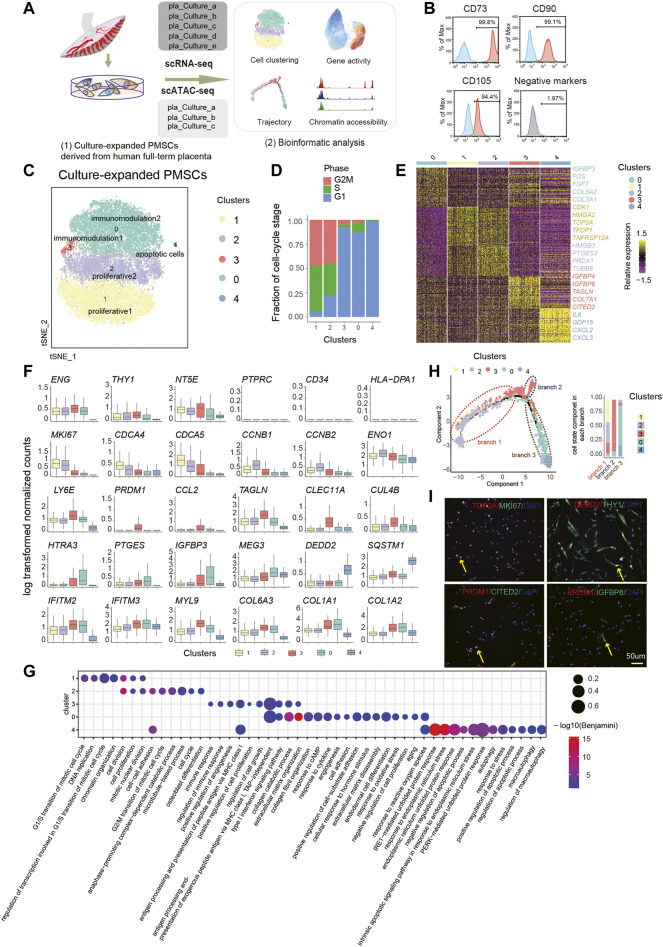
FIGURE 1.
Single-Cell Transcriptome Analysis of PMSC. (A) Schematic overview of the workflow. MSCs derived from human placenta were processed for scRNA-seq (n = 5) and scATAC-seq (n = 3). (B) Representative flow cytometric histogram of PMSCs showing the presence of positive MSC markers (CD73, CD90 and CD105) and absence of negative MSC markers (CD34, CD45, CD11b, CD19, and HLA-DR were merged) (C) t-SNE visualization of 31,219 PMSC cells from five samples reveals heterogeneous cell states at the single-cell RNA seq level. Each dot represents a single cell (n=31,219), colored by its corresponding cluster. (D) Bar plot showing the fraction of cell cycle component in each cluster (bottom). (E) A heat map shows genes (rows) that are differentially expressed across five clusters, colored by relative gene expression (z-score). Gold: high expression; Purple: low expression. Representative genes are highlighted (p<.05, logFC>0.25, top90 in each cluster). (F) Boxplot showing the expression level of selected representative DEGs in five clusters. (G) GO terms enrichment of DEGs respective to indicated PMSC clusters. (H) Pseudotemporal developmental trajectory of PMSCs inferred by Monocle. Bar plot showing the fraction of each cluster component in each branch (bottom). (I) Immunostaining of MKI67, TOP2A, DEDD2, THY1(CD90), PRDM1, CITED2, and IGFBP6 in PMSCs.