Figure 3. The ribosomal DNA damage seen in spinal muscular atrophy is R-loop mediated.
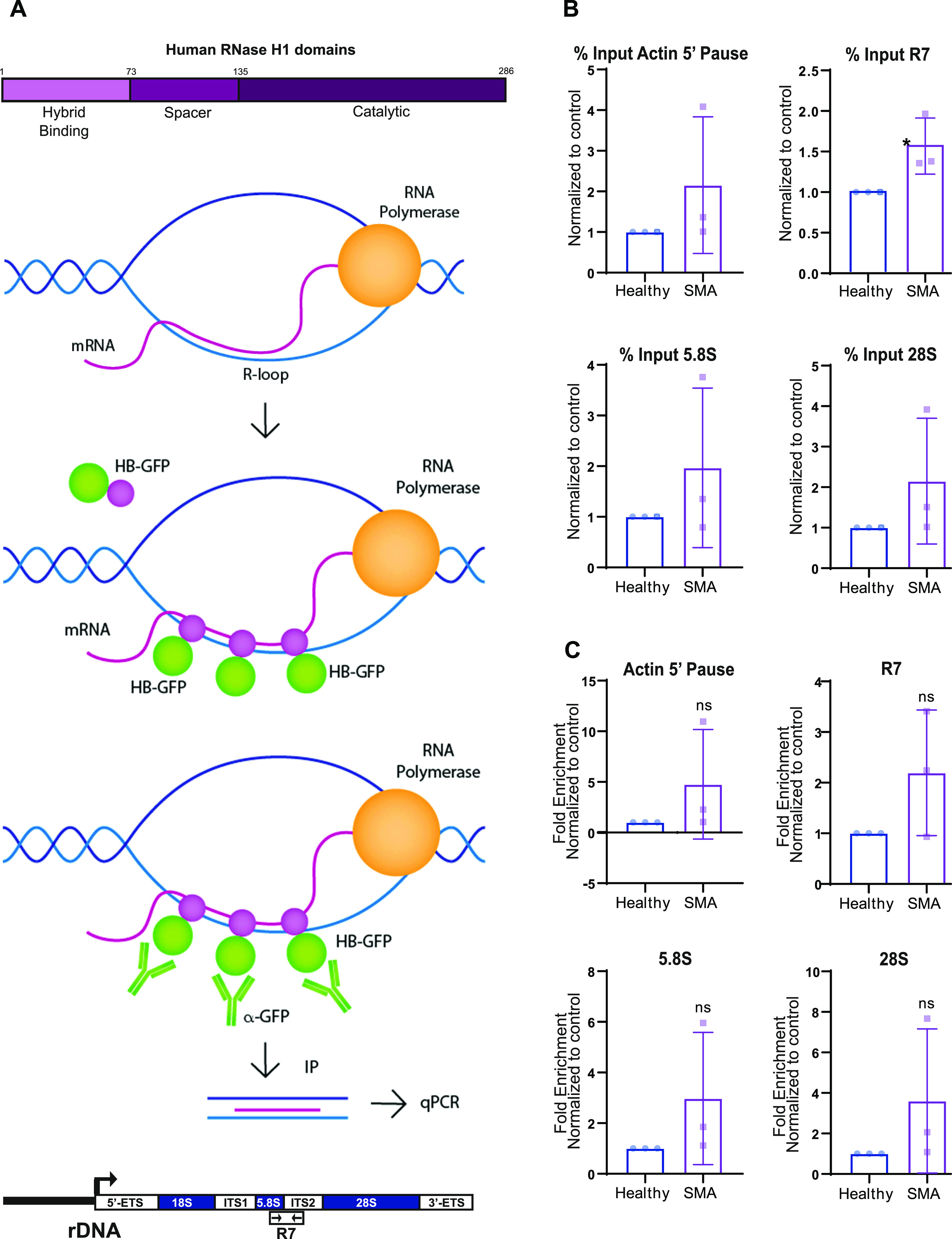
(A) Schematic representation of the HB-GFP-ChIP approach. Hybrid binding domain from RNase H1 fused with EGFP detected and bound to DNA/RNA hybrids. R-loops were isolated by pulling down the HB-GFP fusion protein with GFP-trap beads. qPCR-amplified Actin 5′ pause nuclear region; and R7, 5.8S, and 28S ribosomal sequences. (B, C) Quantified qPCR data from HB-GFP-ChIP experiment in spinal muscular atrophy type I fibroblasts and healthy controls. Data are presented as mean ± s.d. (N = 3). *P < 0.05, ns, not significant (P > 0.05). (B) Percent input, unpaired two-tailed t test; P = 0.3004 (Actin 5′ pause), P = 0.0464 (R7), P = 0.3481 (5.8S), P = 0.2691 (28S). (C) Fold enrichment, unpaired two tailed t test; P = 0.2947 (Actin 5′ pause), P = 0.1702 (R7), P = 0.2603 (5.8S), P = 0.2734 (28S).
