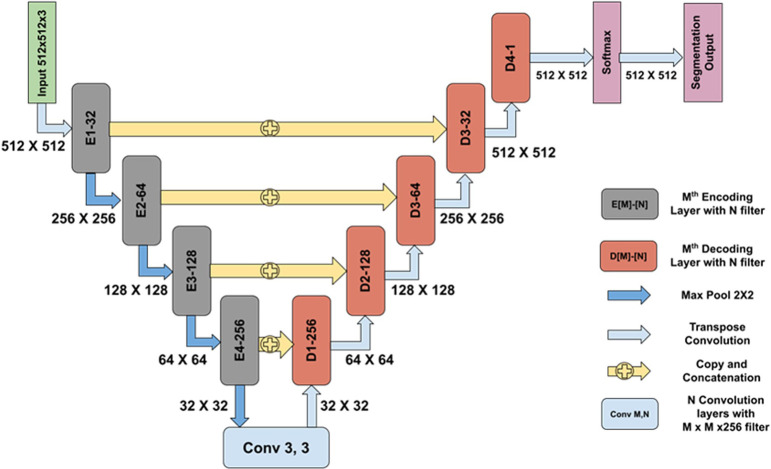
Fig. 2.
Network architecture used for segmentation of various parameters. An encoder–decoder convolutional neural network (CNN) was trained to segment the lesions using annotated training datasets. The network architecture is similar to the U-Net architecture, which has skip connections from the encoding layers to the decoding layers. This helps in eliminating the vanishing gradient problem and thereby simplifies the optimization during the backpropagation of the gradients. In the architecture, E [M]-[N] denotes the Mth encoding layer with N convolutional filters incorporating an inception-like module, whereas D [M]-[N] denotes the Mth decoding layer with N convolutional filters followed by a transposed convolution. Inception modules possess the ability to extract features at multiple scales by employing convolution filters of varying sizes. After each encoding block, the feature map is downsampled by a factor of 2 using a max pool operation on a 2 × 2 receptive field with a stride of 2. This reduces the number of learnable parameters, which in turn reduces the computational cost. At each decoder block (D[M]-[N]), outputs from both the previous decoding layer and the encoding layer are concatenated; they are further convolved with convolution filters followed by a transposed convolution of 2 × 2 with a stride of 2. Unlike the plane upsampling layer, a transposed convolution has learnable parameters that help in the better reconstruction of a segmentation map. Each decoding block increases the feature map size by a factor of 2. A softmax layer is used after the last decoding block to obtain a probability map that determines the probability of each pixel belonging to each class. An argmax operation is performed in the last layer to obtain the segmentation output.