Summary
Aberrant cellular bioenergetics has detrimental consequences in host cells. For instance, pathogenic Zika virus strains can suppress mitochondria respiration and glycolytic functions, disrupting cellular bioenergetics that leads to apoptosis. Herein, we describe methods for flavivirus propagation, titering and infection, cell preparation, and procedures for mitochondrial and glycolytic stress tests. The protocol enables assessment of cellular respiration and glycolytic flux in flavivirus-infected cells.
For complete details on the use and execution of this protocol, please refer to Yau et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Metabolism, Microbiology, Molecular Biology
Graphical abstract

Highlights
-
•
Step-by-step protocol for assessing cellular bioenergetics in flavivirus-infected cells
-
•
Mitochondrial stress test measures respiratory parameters in flavivirus-infected cells
-
•
Glycolysis stress test measures glycolysis parameters in flavivirus-infected cells
Aberrant cellular bioenergetics has detrimental consequences in host cells. For instance, pathogenic Zika virus strains can suppress mitochondria respiration and glycolytic functions, disrupting cellular bioenergetics that leads to apoptosis. Herein, we describe methods for flavivirus propagation, titering and infection, cell preparation, and procedures for mitochondrial and glycolytic stress tests. The protocol enables assessment of cellular respiration and glycolytic flux in flavivirus-infected cells.
Before you begin
Overview
Extracellular Flux (XF) analysis can monitor the oxygen consumption rate (OCR) and the release of hydrogen ions in the extracellular medium (extracellular acidification rate, ECAR) in real time. Measurement of both parameters is fundamental in the understanding of overall cellular bioenergetics as cells primarily utilize oxidative phosphorylation and glycolysis for ATP production. The protocols below describe the specific steps that are involved in assessing rates of mitochondria respiration and glycolysis in MRC-5 cells. We have specifically chosen MRC-5 cells for our assays as these are diploid primary cells, thus minimizing the potentially confounding Warburg effect. However, we have also used this protocol for other adherent cell lines such as HEK293 cells and A549 cells.
Protocol for propagation and titering of Zika and dengue viruses
Timing: 2 weeks
-
1.Virus propagation in Vero cells.
-
a.Seed Vero cells in suitably sized tissue culture vessel, such as T75 flasks, in DMEM growth medium supplemented with 8% FBS (DMEM GM) to a confluency of 80–90% in a 37°C incubator with 5% CO2.
-
b.Decant or aspirate the media from flasks with a serological pipette, and rinse the monolayer of cells once with PBS.
-
c.Infect the cells with multiplicity of infection (MOI) 0.1 Zika or dengue virus diluted in DMEM maintenance medium supplemented with 2% FBS (DMEM MM) for 1 h in 37°C incubator with 5% CO2. Gently rock the flasks from side to side, every 15 min, to ensure good contact between the virus and cells, and to prevent cells from drying out.
-
d.After 1 h absorption, discard the virus inoculum and replace with fresh DMEM MM.
-
e.Incubate the cells in 37°C incubator with 5% CO2 for 5–7 days. There is no need to change the media at this step.
-
f.Harvest the virus supernatant when 70–90% cytopathic effect can be observed.
-
g.Clarify the virus supernatant by centrifuging at 1,200 rpm for 4 min (288 × g) to remove cellular debris and passing through a 0.22 μM filter.
-
h.Aliquot and store the virus in −80°C freezer. Quantify infectious titer of viruses using plaque assay as detailed below.
-
a.
-
2.Plaque assay.
-
a.Seed about 1 × 105 cells Vero cells in 24-well tissue culture plates in DMEM GM in a biosafety cabinet.
-
b.Incubate cells in a 37°C incubator with 5% CO2 for 1 day, until a nice monolayer of cells can be seen.
-
c.Thaw an aliquot of the virus cultured above and perform 10-fold serial dilutions in DMEM MM. We recommend doing at least 6 serial dilutions.
-
d.Aspirate and discard the cell media from wells.
-
e.Infect the cells with 100 μL/well of the serially diluted virus in duplicates. Incubate for 1 h in 37°C incubator with 5% CO2. Gently rock the plates from side to side, every 15 min, to ensure good contact between the virus and cells, and to prevent cells from drying out.
-
f.After 1 h absorption, discard the virus inoculum and replace with DMEM carboxymethyl cellulose overlay medium.
-
g.Incubate plates for 6 days in 37°C incubator with 5% CO2,. There is no need to change the media at this step.
-
h.Incubate the plates with 10% formalin for 30 min to fix the cells and inactivate the viruses.
-
i.Discard the supernatant by decanting or aspirating with a serological pipette. Rinse plate under running tap water.
-
j.Stain the cell monolayer with 1% crystal violet for approximately 15–30 min.
-
k.Wash the plate extensively under running tap water to visualize the plaques.
-
l.Allow the plates to air dry before counting the number of plaques to determine the virus titer in plaque forming unit per milliliter (pfu/mL) (Figure 1).
-
a.
Note: If working with infectious clones, it is recommended that viruses are not passaged more than twice to minimize chances of virus mutation. It is also important to sequence the entire virus genome to ensure that virus sequences have not mutated in cell culture prior to experiment.
Note: All viruses used for Extracellular Flux assays will need to be at least 106 pfu/mL.
Figure 1.
Example of a plaque assay result after crystal violet staining
Vero cells are fixed with formalin and stained with crystal violet after infection with H/PF/2013. Clear areas (plaques) indicate presence of viruses infecting the cells. In this figure, 10-fold serial dilutions of the viruses are performed in replicates, starting from 1000-fold dilution on the first column. Plaque assay performed in the 24-well plate format.
Subculture protocol for MRC-5 cells
Timing: Subculture once every 3–4 days
The following protocol is suitable for cells cultures in T75 flasks. MRC-5 are passaged using a 1:2 split ratio.
-
3.
Check the condition of the cells prior to passage.
-
4.
Warm up Eagle’s Minimum essential Medium (EMEM) growth medium supplemented with 8% FBS (GM) and trypsin in a 37°C bath for at least 30 min.
-
5.
Discard the growth media by either decanting or aspirating with a serological pipette.
-
6.
Wash the cells by adding 2 mL of 1× PBS. Discard by decanting or aspirating with a serological pipette.
-
7.
Add 2 mL of trypsin into the flask and incubate in 37°C for 2 min.
-
8.After 2 min, gently tap the sides of the flask to dislodge the cells.
-
a.You can visualize the cells under a microscope to ensure that most of the cells have been dislodged.
-
a.
-
9.
Add 2 mL of EMEM GM into each T75 flask.
-
10.
Harvest and collate all the cells in a 15 mL tube.
-
11.
Spin down at 1,200 rpm (288 × g) for 4 min at 20°C–25°C.
-
12.
Add 9 mL fresh culture media into each new T75 flask.
-
13.
After spinning down, discard or aspirate the media and dislodge the cell pellet via gentle flicking.
-
14.For every T75 flask prepared, add 2 mL of fresh culture media into the 15 mL tube. Thereafter, add 1 mL of fresh culture media for every T75 flask prepared. This will ensure that the cells will be split at a 1:2 split ratio.
-
a.Gently rock the flask a few times and let the cells settle.
-
a.
-
15.
Incubate the cells at 37°C at 5% CO2. Flasks should be confluent after 2 days.
Note: A minimum of 2.5 × 106 MRC-5 cells in total is required for seeding one day prior to infection. Cells should be maintained at low passage, preferably less than 28 passages as cellular senescence will start to occur after 35 passages.
Note: If T75 flasks are not readily available, users may consider using petri dishes to culture the cells. Users can use 2 mL of EMEM GM to wash the cells and 10 mL of new media to culture the cells, at a split ratio of 1:2. The seeding density for a 60 mm dish is approximately 1 × 106 cells.
Note: For A549 and HEK293, we recommend using DMEM GM to culture the cells. A split ratio of 1:5 and 1:8 is suitable for culturing these cells respectively.
Note: Routine test for Mycoplasma, bacteria and yeast should be carried out to ensure that the cells used for experiments are not contaminated with another infection.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Cell lines | ||
| Vero cells | ATCC | Cat# CCL-81 |
| MRC-5 cells | ATCC | Cat# CCL-171 |
| HEK293 cells | ATCC | Cat# CRL-1573 |
| A549 cells | ATCC | Cat# CRM-CCL-185 |
| Bacterial and virus strains | ||
| Zika MR766, GENBANK: M143022.1 | ATCC | Cat #VR-1838 |
| Zika H/PF/2013, GENBANK: KJ776791.2 | European Virus Archive | N/A |
| Zika DN-2, Virus made from PF13/251013-18 (GENBANK: KX369547) as viral backbone | Derived from infectious clone (Kwek et al., 2018) | N/A |
| Zika DN-1, Virus made from PF13/251013-18 (GENBANK: KX369547) as viral backbone | Derived from infectious clone (Kwek et al., 2018) | N/A |
| Zika M-F37L, Virus made from PF13/251013-18 (GENBANK: KX369547) as viral backbone | Derived from infectious clone (Yau et al., 2021) | N/A |
| Dengue virus | Isolated from patient (Low et al., 2006) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM with L-glutamine and Sodium Pyruvate | Gibco, Thermo Fisher Scientific | Cat# 11995 |
| Fetal Bovine Serum (FBS) | HyClone | Cat# SH30396.03 |
| 1 M Hepes | Lonza | Cat# 17-737E |
| Penicillin/Streptomycin (Pen/Strep) | HyClone | Cat# SV30010 |
| MEM with L-glutamine | Gibco, Thermo Fisher Scientific | Cat# 11095 |
| L-glutamine (100×) | Gibco, Thermo Fisher Scientific | Cat# 25030081 |
| DMEM, powder, high glucose | Gibco, Thermo Fisher Scientific | Cat# 12100046 |
| Aquacide II (Carboxymethyl cellulose) | Merck Millipore | Cat# 17851 |
| Trypsin-EDTA (0.25%), Phenol Red | Gibco, Thermo Fisher Scientific | Cat# 25200072 |
| Trypan Blue | Sigma-Aldrich | Cat# 15250061 |
| Crystal Violet Solution | Sigma-Aldrich | Cat# V5265 |
| 10× PBS | Sigma-Aldrich | Cat# BUF-2040-10X4L |
| 0.22 μM filter | Nalgene | Cat# 595-4520 |
| Oligomycin A | Sigma-Aldrich | Cat# 209-437-3 |
| Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) | Sigma-Aldrich | Cat# 206-730-8 |
| Rotenone | Sigma-Aldrich | Cat# 83-79-4 |
| Antimycin A from Streptomyces sp. | Sigma-Aldrich | Cat# A8674 |
| Glucose | Sigma-Aldrich | Cat# G7021 |
| Sodium pyruvate | Sigma-Aldrich | Cat# 5280 |
| Glutamine | Sigma-Aldrich | Cat# G8540 |
| 2-deoxy-D-glucose | Sigma-Aldrich | Cat# D8375 |
| Critical commercial assays | ||
| Seahorse XF24 V28 PS Cell Culture Microplates | Agilent | Cat# 100882-004 |
| Seahorse XF24 Islet Capture Microplates | Agilent | Cat# 101122-100 |
| Seahorse XF RPMI Medium, pH 7.4, 500 mL | Agilent | Cat# 103576-100 |
| Seahorse XF Calibrant Solution | Agilent | 100 mL (Cat# 103059-000) or 500 mL (Cat# 100840-000) |
| Seahorse XFe24 Analyzer | Agilent | N/A |
| Software and algorithms | ||
| Seahorse Wave Desktop Software | Agilent | N/A |
Alternatives: Users can also consider using the Seahorse XF Glucose (1.0 M solution) (Cat #103577-100), Seahorse XF Pyruvate (100 mM solution) (Cat #103578-100) and Seahorse XF Glutamine (200 mM solution) (Cat #103579-100) from Agilent for the Seahorse experiments.
Materials and equipment
DMEM Growth Media (DMEM GM)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM with L-glutamine and sodium pyruvate | 91% | 455 mL |
| Heat inactivated FBS | 8% | 40 mL |
| Hepes | 1% | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 1 month.
DMEM Maintenance Media (DMEM GM)
| Reagent | Final concentration | Final concentration |
|---|---|---|
| DMEM with L-glutamine and sodium pyruvate | 96% | 480 mL |
| Heat inactivated FBS | 2% | 10 mL |
| Hepes | 1% | 5 mL |
| Penicillin/Streptomycin | 1% | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 1 month.
DMEM carboxymethyl cellulose overlay medium
Mix equal volumes of 2× DMEM media with 1.6% carboxymethyl cellulose solution to make DMEM carboxymethyl cellulose overlay medium. Store at 4°C for up to 1 month.
The reagents and procedures to make 2× DMEM media and 1.6% carboxymethyl cellulose solution are:
2× DMEM media
| Reagent | Amount |
|---|---|
| DMEM, powder, high glucose | 1 pack |
| Distilled water | 470 mL |
| Heat inactivated FBS | 20 mL |
| Hepes | 10 mL |
| Penicillin/Streptomycin | 10 mL |
| Total | 500 mL |
Sterilize by passing media through the 0.2 μm filter. Store at 4°C for up to 1 month.
1.6% carboxymethyl cellulose solution
| Reagent | Amount |
|---|---|
| Carboxymethyl cellulose | 8 g |
| Distilled water | 500 mL |
Autoclave at 121°C for 20 min to sterilize the solution. Store at 4°C for up to 1 month.
Preparation of Seahorse assay media
Prepare the Seahorse XF RPMI assay media or Seahorse glycolysis assay media using the media compositions as stated below. The media should preferably be prepared on the day of the seahorse experiment. Details on the composition of the assay media are described below:
Seahorse XF RPMI Assay Media
| Reagent | Concentration |
|---|---|
| Seahorse XF RPMI base media | – |
| Glucose | 25 mM |
| Pyruvate | 1 mM |
| Glutamine | 2 mM |
Adjust pH to 7.4 after adding all reagents. Prepare when required and use immediately.
Seahorse Glycolysis Assay Media
| Reagent | Concentration |
|---|---|
| Seahorse XF RPMI base media | – |
| Glutamine | 2 mM |
Adjust pH to 7.4 after adding all reagents. Prepare when required and use immediately.
Store all stock assay media and at 4°C for up to a year, protected from light. Ensure that the XF media are within expiry date.
Note: When preparing Seahorse XF RPMI assay media for mitochondria stress test or glycolytic stress test, glucose, pyruvate and glutamine can be dissolved or diluted in 1× sterile PBS.
Preparation of drugs
| Test | Reagent | Stock concentration |
|---|---|---|
| Mitochondrial stress test | Oligomycin (10,000×) | 10 mM |
| FCCP (10,000×) | 15 mM | |
| Antimycin-A (10,000×) | 10 mM | |
| Rotenone (10,000×) | 1 mM | |
| Glycolysis stress test | 2-DG (1,000×) | 1 M |
| Glucose (1,000×) | 1 M |
Store drugs at −20°C for up to one year. Avoid freeze-thaw cycles to retain drug activity.
Note: Users can dilute the drugs with DMSO to the appropriate concentrations as indicated above. Otherwise, users can use the drugs directly from either the Mito Stress test kit (Cat #103015-100) or the Glycolysis Stress test kit (Cat #103020-100).
Note: Drugs are prepared at 1,000× - 10,000× concentrations for ease of use on actual day of experiment. Store drugs at −20°C.
Note: CCCP and Piericidin A can be potentially used instead of FCCP and rotenone respectively. However, users will need to test the drug concentrations to make sure that they work optimally.
Step-by-step method details
Cell seeding protocol on XF cell culture plates
Timing: 1 day
The protocol is designed to allow sufficient time for MRC-5 cells to adhere onto the XF cell culture plates for XF analysis. The wells should be about 90% confluent after 18–24 h incubation.
Day 1.
-
1.
Culture an appropriate number of cells (minimum 2.4 × 106 cells) for your experiment. Refer to the “Subculture protocol for MRC-5 cells” described in the earlier section for more information.
-
2.Seed MRC-5 cells in Seahorse XF 24-well plates 24 h before the optimization test or study protocol.
-
a.Dilute cells to 2 × 105 cells/mL using the EMEM GM.
-
b.Add 500 μL of cell suspension into each well, amounting to 1 × 105 cells/well.
-
a.
-
3.
Tap the sides of the plate gently and let the cells attach in 37°C, 5% CO2 for 18–24 h.
Day 2.
-
4.Check viability of cells prior to the start of the experiment. The cells can be visualized using a conventional light microscope.
-
a.Ensure a confluent (>90%) monolayer of cells have been formed. There should be minimal well to well variability as this will affect the quality of your results.
-
a.
-
5.
The cells are now ready for the optimization or test protocol.
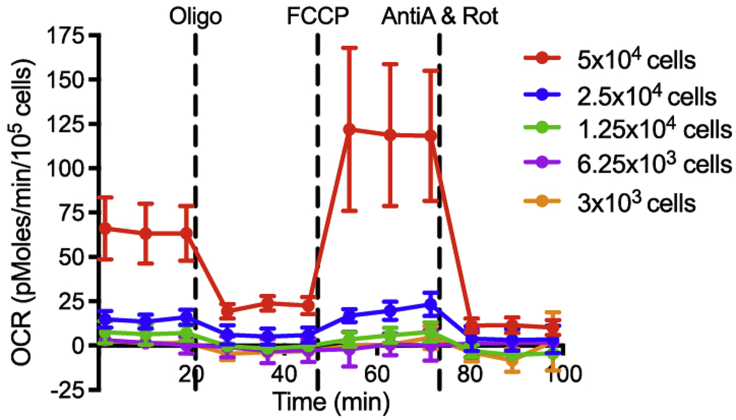
CRITICAL: Different cell types have different metabolic activity. If the XF analysis is performed for another cell type, we strongly recommend optimizing the cell numbers by doing an XF analysis on different cell counts. We find that in most cases, using 1 x 105 cells gave the most robust signal. An example of different MRC-5 cell numbers on the mitochondria stress profile is provided (Figure 2). Note that a minimum of 5 × 104 cells are required for OCR signals to be detected, which is the reason why we have chosen to use 1 × 105 MRC-5 cells for XF analysis.
Note: We have also optimized the cell densities for A549 and HEK293 cells, where 1 × 105 cells are suitable for XF analysis.
Note: We have used the 24-well plates for all our experiments as we are able to obtain consistent and reproducible read-outs in our experimental replicates. However, if you want to perform these experiments on a 96-well format, you will have to adapt the volumes and cell numbers accordingly (van der Windt et al., 2016). Users may also refer to the Agilent website at https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay for more details.
Note: If you need to perform pre-treatment of cells before virus infection, you may incubate the chemical or drug with the cells during or after cell seeding.
Figure 2.
Mitochondrial respiration profile for MRC-5 cells
Different numbers of MRC-5 cells are subjected to the mitochondrial stress test. Results show that a minimum of 5 × 104 cells are required for OCR signals to be detected. Data are displayed as mean ± SD.
Virus infection assay protocol on XF cell culture plates
Timing: 2 days
After cell seeding and incubation for 18–24 h, the cells are now ready to be infected. This protocol describes the steps involved in infecting MRC-5 cells seeded on 24-well XF Cell Culture Microplate.
-
6.Thaw the viruses in a water bath at 37°C and dilute the viruses to the MOI 1.0 using EMEM maintenance media (MM). Keep the virus on ice when not in use.
-
a.We recommend 4 replicates for each experimental condition, including negative controls and at least one blank well containing only media. The negative controls should be composed of cells without virus and drug treatment. One plate hence allows testing of up to 6 different experimental conditions.
-
b.Dilute viruses to 1 × 107 pfu/mL with EMEM MM.
-
a.
-
7.Infect MRC-5 at MOI 1.
-
a.Remove 350 μL of EMEM GM and add 100 μL of pre-diluted virus into each well.
-
b.Allow infection to proceed for 1 h in 37°C incubator, 5% CO2. Rock the plate once every 15 min.
-
a.
-
8.After 1 h, remove the virus using serial dilution. This is to prevent the well from drying.
-
a.Add 250 μL of EMEM MM into each well.
-
b.Remove 400 μL of media and replace with a similar volume. Repeat this step 3 times.
-
a.
-
9.
Incubate the infected cells for 48 h at 37°C, 5% CO2.
-
10.
After 48 h, proceed with the extracellular flux analysis (either mitochondrial stress test or glycolytic stress test).
Note: Ensure that the viruses are taken out of the water bath immediately after thawing to prevent virus inactivation.
Note: It is important to optimize the multiplicity of infection and the duration of infection to ensure that there are no extensive cytopathic effects during the day of XF analysis. For MRC-5, extensive cytopathic effects were noted at 65–72 hours post infection.
Note: We recommend having at least 4 biological replicates for each experimental condition, although it is theoretically possible to perform them in triplicates. In addition, a negative control without virus and drug treatment should be included, so that we can compare the metabolism profiles between the infected and uninfected conditions. At least one blank well containing only media will also be needed to determine the background signal.
Note: Media should be added or removed carefully to prevent dislodging the adherent cells. We recommend adding or removing media by placing the pipette tip at the side of the walls to reduce turbulence.
Note: The duration of infection may vary depending on the virus and cell types used. Ensure that the cytopathic effects are not visible during the assessment of extracellular flux, as reduced cell numbers can affect the results and interpretation of the extracellular flux analysis.
Extracellular flux analysis to monitor mitochondrial oxygen consumption rate
Timing: 2 days
Timing: 4–5 h
Mitochondria is the powerhouse of the cell that generates ATP through oxidative phosphorylation. The mitochondrial respiration test utilizes the oxygen consumption rate (OCR) as a direct read-out for cell respiration rate. The key respiratory parameters that can be measured are: (i) basal respiration, which is the initial OCR minus the non-mitochondrial respiration, (ii) ATP-linked OCR, which is determined after addition of oligomycin that inhibits ATP synthase, (iii) proton leak, which is the difference between ATP-linked OCR and non-mitochondrial respiration, (iv) maximal respiration, which is induced after addition of carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) that is a potent mitochondrial oxidative phosphorylation uncoupler, (v) spare respiratory capacity, which is the difference between maximal respiration and basal respiration and (vi) non-mitochondrial respiration, which is the OCR after addition of rotenone (complex I inhibitor) and antimycin A (complex III inhibitor) (Figure 3). The protocol hence describes the steps required to prepare the drugs and cells for mitochondrial stress test.
Figure 3.
Measurements of the different parameters from the mitochondrial respiration test
Schematic showing the different respiratory parameters that can be measured by the mitochondrial respiration test. Data are displayed as mean ± SD.
Day prior to mitochondria stress test seahorse experiment.
-
11.
At one day before XF analysis, hydrate a sensor cartridge and utility plate for 18–24 h, with 1 mL of XF Calibrant in a non-CO2 37°C incubator. Ensure that the incubator is properly humidified. Users can refer to https://www.agilent.com/en/product/cell-analysis/how-to-run-an-assay for specific instructions on the hydration protocol to use for the different cartridges. Users will have to perform hydration before performing the mitochondrial respiration test or glycolytic stress test.
Day of assay.
-
12.Dilute the drugs from their respective stock concentrations to 10× in Seahorse XF RPMI assay media beforehand.
-
a.Refer to the materials and methods segment for detailed instructions on how to prepare the Seahorse XF RPMI assay media.
-
b.Dilute drugs to 10× to a total volume of 3 mL using the following dilution ratios:
-
i.Oligomycin: Add 30 μL (10,000×) to 2,970 μL of Seahorse XF RPMI assay media.
-
ii.FCCP: Add 30 μL (10,000×) to 2,970 μL of Seahorse XF RPMI assay media.
-
iii.Rotenone + antimycin A: Add 30 μL of rotenone (10,000×) and 30 μL of antimycin A (10,000×) to 2,940 μL of Seahorse XF RPMI assay media.
-
i.
-
a.
-
13.
Wash cells with Seahorse XF RPMI assay media. Remove 300 μL of media and replace with 300 μL of Seahorse XF RPMI assay media for each wash. Repeat the washes for a total of three times.
-
14.
After the last wash, top up each well using Seahorse XF RPMI assay media to a final volume of 530 μL.
-
15.
Observe cells under the light microscope to ensure that a monolayer of cells is present in all wells, and that no cells were dislodged.
-
16.
Let the cell equilibrate in the Seahorse XF RPMI assay media for at least 30 min. The plate can be placed in a humidified chamber during transit to a facility. If not, the plate can be placed in a regular incubator at 37°C with 5% CO2.
-
17.Pipet the drugs into the drug ejection ports with the volumes as follows:
-
a.Port A: Oligomycin (70 μL).
-
b.Port B: FCCP (75 μL).
-
c.Port C: Rotenone + Antimycin A (85 μL).
-
a.
-
18.Switch on the Seahorse XF analyser. In the “Wave” software, select the mitochondria stress test assay template, which will assign ports A, B and C to be oligomycin, FCCP and rotenone + antimycin A respectively. After which, click “Design” to assign the sample positions according to the experimental conditions. A standard seahorse run includes the following steps:
-
a.Calibrate.
-
b.Equilibrate.
-
c.Baseline readings: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
d.Inject port A: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
e.Inject port B: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
f.Inject port C: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
g.End program.
-
a.
-
19.
When prompted, place the loaded sensor cartridge with the utility plate into the Seahorse analyser and start the calibration step.
-
20.
After the calibration step, replace the calibrant plate for the cell plate. Ensure that the cell plate is loaded in the correct orientation.
-
21.
Allow the seahorse experiment to run as per preset protocol.
-
22.
Extract the results for analysis with the Seahorse Wave Desktop Software. Users can download the software at https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897. The step-by-step protocol on how to use the software can be located at https://hpst.cz/sites/default/files/oldfiles/user-guide-wave-desktop-22.pdf.
-
23.
Dispose of all consumables as biohazardous waste.
Note: Make sure that the sensor cartridge and utility plate are hydrated one day before running the XF analysis.
Note: Wash the cells gently to minimize the risk of cells lifting off. It is often a good habit to place the pipet tip against the side of the walls before dispensing to minimize disruption of the monolayer. Check that the cells remain intact after the washing steps.
Note: While it is theoretically possible to perform the mitochondria stress test and glycolytic stress test simultaneously within a single plate, we do not recommend doing so as the media for cell washing and drugs that are used for both tests are different. If both assays are concurrently running in one plate, ensure that the ports A, B and C are loaded with the correct drugs.
Note: The activity of FCCP may decline upon multiple freeze-thaw cycles. We recommend keeping FCCP in small aliquots to preserve the FCCP activity for longer periods of time.
Note: You can collect the supernatant before performing the cell washes and perform a plaque assay to ascertain the level of virus infection during the point of XF analysis.
Note: Normalization is critical in the workflow for analysis of raw data from the XF analyzer. In this protocol we used the number of cells as the method for normalization. However, other methods such as protein concentration and DNA content can be used as well. For details, users can refer to https://www.agilent.com/cs/library/technicaloverviews/public/Methods_and_Strategies_for_Normalizing_Tech_Overview_022118.pdf.
Extracellular flux analysis to monitor glycolytic rates
Besides oxidative phosphorylation, glycolysis is the second pathway that supplies ATP to satisfy the energy requirements of cells. This protocol measures the different glycolytic parameters after virus infection, where the extracellular acidification rate (ECAR) is predominantly the read-out for excretion of lactic acid per unit time. The key parameters measured are: (i) glucose metabolism, which is the difference between maximum ECAR measurement after glucose addition and non-glycolytic acidification, (ii) maximal glycolytic capacity, which is the difference between maximum ECAR measurement after inhibition of oxidative phosphorylation by oligomycin and non-glycolytic acidification, (iii) spare glycolytic reserve, which is the difference between maximal glycolytic capacity and glucose metabolism, and (iv) non-glycolytic extracellular acidification rate which is the glucose metabolism rate after treatment with 2-deoxyglucose, which is an inhibitor of hexokinase II that catalyzes the first step of glycolysis (Figure 4). This protocol describes the steps required to prepare the drugs and cells for glycolytic stress test. The results of the glycolytic stress test will complement with the mitochondrial stress test, providing a comprehensive overview of the cellular bioenergetics involved.
-
24.
Conduct steps 1–25 as detailed above.
Figure 4.
Measurements of the different parameters from the glycolytic stress test
Schematic showing the different glycolytic parameters that can be measured by the glycolytic stress test. Data are displayed as mean ± SD.
Day prior to glycolytic stress test seahorse experiment.
-
25.
At one day before XF analysis, hydrate a sensor cartridge and utility plate for 18–24 h, in a non-CO2 37°C incubator. Ensure that the incubator is properly humidified.
On the day of assay.
-
26.Dilute the drugs to 10× in Seahorse glycolysis assay media.
-
a.Refer to the materials and methods segment for detailed instructions on how to prepare the Seahorse glycolysis assay media.
-
b.Dilute drugs from 1,000× to 10× with a total volume of 3 mL for each drug, using the following dilution ratios:
-
i.Port A: Glucose (Add 30 μL–2,970 μL of Seahorse glycolysis assay media).
-
ii.Port B: Oligomycin (Add 30 μL–2,970 μL of Seahorse glycolysis assay media).
-
iii.Port C: 2-DG (Add 30 μL–2,970 μL of Seahorse glycolysis assay media).
-
i.
-
a.
-
27.
Wash cells with Seahorse glycolysis assay media. Remove 300 μL of media and replace with 300 μL of Seahorse glycolysis assay media for each wash. Repeat the washes for a total of three times.
-
28.
After the last wash, top up each well using Seahorse glycolysis assay media to a final volume of 530 μL.
-
29.
Observe cells under the light microscope to ensure that a monolayer of cells is present in all wells, and that no cells were dislodged.
-
30.Pipet the drugs into the drug ejection ports with the volumes as follows:
-
a.Port A: Glucose (70 μL).
-
b.Port B: Oligomycin (75 μL).
-
c.Port C: 2-DG (85 μL).
-
a.
-
31.Switch on the Seahorse XF analyzer. In the “Wave” software, select the glycolytic rate assay template, which will assign ports A, B and C to be oligomycin, FCCP and rotenone + antimycin A respectively. After which, click “Design” to assign the sample positions according to the experimental conditions. A standard seahorse run includes the following steps:
-
a.Calibrate.
-
b.Equilibrate.
-
c.Baseline readings: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
d.Inject port A: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
e.Inject port B: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
f.Inject port C: Loop 3 times of mix (3 min), wait (2 min) and measure (3 min).
-
g.End program.
-
a.
-
32.
When prompted, place the loaded sensor cartridge with the utility plate into the Seahorse analyzer and start the calibration step.
-
33.
After the calibration step, replace the calibrant plate for the cell plate. Ensure that the cell plate is loaded in the correct orientation.
-
34.
Allow the seahorse experiment to run as per preset protocol.
Extract the results for analysis with the Seahorse Wave Desktop Software. Users can download the software at https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897. The step-by-step protocol on how to use the software can be located at https://hpst.cz/sites/default/files/oldfiles/user-guide-wave-desktop-22.pdf.
-
35.
Dispose off all consumables as biohazardous waste.
Note: Wash the cells gently to minimize the risk of cells lifting off. It is often a good habit to place the pipet tip against the side of the walls before dispensing to minimize disruption of the monolayer. Check that the cells remain intact after the washing steps.
Note: You can collect the supernatant before performing the cell washes and perform a plaque assay to ascertain the level of virus infection during the point of XF analysis.
Note: Normalizations is critical in the workflow for analysis of raw data from the XF analyzer. In this protocol we used the number of cells as the method for normalization. However, other methods such as protein concentration and DNA content can be used as well. For details, users can refer to https://www.agilent.com/cs/library/technicaloverviews/public/Methods_and_Strategies_for_Normalizing_Tech_Overview_022118.pdf.
Expected outcomes
To ascertain that the experiments are working as intended, the uninfected cells should display a typical “Seahorse” shape for the respiration profile. The addition of oligomycin, which inhibits ATPase should significantly reduce OCR readings. Thereafter, the addition of FCCP, which acts as a proton ionophore, should result in maximal respiration. Finally, the addition of antimycin and rotenone, which inhibits complex III and complex I activity, should then disrupt all mitochondrial activity, resulting in minimal respiration (Figure 5A). In our illustrated example, we demonstrated that pathogenic Zika virus strains (M-F37L, H/PF/2013, MR766) resulted in significantly reduced maximal respiration and spare respiratory capacity whereas the attenuated Zika virus strains (DN1, DN2) did not affect respiratory profiles (Figures 5A and 5B).
Figure 5.
Respiration and glycolytic profiles of MRC-5 cells infected with different Zika virus strains
Pathogenic Zika virus strains (M-F37L, MR766 and H/PF/2013) infection results in significant reduction in maximum respiration, spare respiratory capacity (SRC) and glycolytic capacity.
(A) Respiratory profiles (OCR) of MRC-5 cells after 48 h post-infection with DN-2 (blue), DN-1 (light blue), M-F37L (purple), MR766 (orange), and H/PF/2013 (red). After basal respiration, cells were subjected tooligomycin (oligo), which inhibits ATP synthase; followed by FCCP which uncouples mitochondrial respiration and maximizes OCR; and finally, anti-mycin A and rotenone (AntiA and Rot), which inhibit complex III and I, respectively.
(B) Measurements of the different respiratory parameters under different infection conditions. Values derived from panel (A).
(C) Extracellular acidification profile (ECAR) of MRC-5 cells after 48 h post-infection with various ZIKV strains. After non-glycolytic acidification, glucose was added to cells, followed by oligomycin to obtainmaximal glycolytic capacity, and finally 2-deoxy-glucose (2-DG) to inhibit glycolysis.
(D) Measurements of the different glycolytic parameters under the different infection conditions. Values derived from panel (C).
For the detailed molecular mechanisms involved, please refer to Yau et al. (2021). Data are displayed as mean ± SD. ∗ p < 0.05, + p < 0.01, # p < 0.001 compared to uninfected cells.
For the glycolysis stress test, the addition of glucose will augment ECAR readings, as glucose will be converted to lactic acid. The addition of oligomycin, which blocks oxidative phosphorylation will further promote ECAR, as the glycolytic substrates will be diverted to lactic acid to generate energy for the cell. Finally, 2-DG which inhibits glycolysis, will then reduce ECAR to baseline levels (Figure 5C). In our illustrated example, we demonstrated that pathogenic Zika virus strains (M-F37L, H/PF/2013, MR766) resulted in significantly reduced glucose metabolism, glycolytic capacity and glycolytic reserve, whereas the attenuated Zika virus strains (DN1, DN2) did not affect the glycolytic profiles (Figures 5C and 5D).
Limitations
The current XF assay measures bulk changes in OCR and ECAR and thus, unable to assess the relative contributions of virus-infected cells and the neighboring uninfected cells to the OCR and ECAR readings. More advanced techniques, such as SCENITH, which uses a flow cytometry platform to profile bioenergetics with single-cell resolution (Arguello et al., 2020) may be potentially employed to evaluate metabolic responses in both infected and uninfected cells.
Troubleshooting
Problem 1
Low signal detection for OCR and ECAR (steps 22 and 34).
Potential solution
Check that cells are cultured in the appropriate media as recommended by ATCC (See key resources table).
Check cell viability using the Trypan Blue dye exclusion test (step 4).
Ensure cells are not contaminated with microorganisms (e.g., Mycoplasma, bacteria, yeast) (steps 1–5).
Ensure cells are not at high passage, as cells tend to undergo cell senescence with increasing cell passage (steps 3–14).
Optimize cell numbers by doing XF analysis on different cell counts (Figure 1).
Ensure cells are gently washed and the washes do not cause substantial loss in cell numbers (steps 13 and 27).
Check the expiry of drugs, particularly for FCCP. Ensure that the FCCP is not previously subjected to multiple freeze-thaw cycles.
Problem 2
Low signal detection for OCR and ECAR for the virus infected cells but not the uninfected cell controls (steps 22 and 34).
Potential solution
Check if the virus has caused extensive cytopathic effects, resulting in substantial cell loss. If that is the case, evaluate for cell apoptosis by measuring for caspase 3 activity, DNA fragmentation assays or annexin V staining. Alternatively, shorten the duration of virus infection or change the multiplicity of infection to better capture the OCR and ECAR readings before massive cell death.
Problem 3
High variance in OCR and ECAR readings between experimental replicates (steps 22 and 34).
Potential solution
Check that the cells are not clumpy, and ensure an equal number of cells are seeded into each well. If needed, users can consider using a 37–70 μm cell strainer to obtain individual cells (step 4).
Ensure similar cell confluency between wells by checking under the light microscope (step 4).
Ensure cell washes are consistent between replicates and the washes do not cause substantial loss in cell numbers (steps 13 and 27).
Check for good experimental techniques to minimize experimental errors.
Increase number of experimental replicates to reduce variation.
Problem 4
Inconsistent respiratory or glycolytic profiles between experiments (steps 22 and 34).
Potential solution
Check that the cell culture media, supplements added to the media and the culturing conditions are similar between experiments (See key resources table).
Ensure that the viruses used in the experiments have not mutated by sequencing the viruses used.
Repeat experiment to validate the observations.
Problem 5
No difference in respiration and glycolysis measurements between infected and uninfected cells (steps 22 and 34).
Potential solution
Check the level of virus infection, preferably by using flow cytometry to ascertain the percentage of cells that are infected at the time-point of XF analysis. If the level of infection is low, increase the multiplicity of infection or increase the duration of infection.
Include a positive control that is shown to affect cellular bioenergetics (e.g., wild-type Zika virus strains can be purchased from ATCC) (See key resources table).
If cells are successfully infected, but cellular bioenergetics remain unchanged, it is possible that the virus does not alter cellular bioenergetics. In our experience, attenuated Zika virus strains (e.g., DN1 and DN2) and wild-type dengue virus strains does not influence cellular bioenergetics in MRC-5 cells, although the cells are clearly infected (Figure 5).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kuan Rong Chan (kuanrong.chan@duke-nus.edu.sg).
Materials availability
All materials are available upon reasonable request.
Acknowledgments
We thank the members of Eng Eong Ooi lab for sharing their observations and difficulties which helped in making the protocols better. We also thank Dr. Yap Lai Lai from the Department of Biochemistry, NUS, for her assistance with the Seahorse experiments. This work is funded by the Individual Research Grant (MOH-000610-00) and the Khoo Pilot Award (Collaborative).
Author contributions
K.R.C., J.Z.H.L., C.Y., S.L.X.Z., and E.E.O. wrote the manuscript. K.R.C., J.Z.H.L., and H.C.T. performed the experiments. K.R.C. and E.E.O. supervised the study.
Declaration of interests
E.E.O. has an issued patent titled “Rapid method of generating live attenuated vaccines” (Singapore patent publication number: 10201602980W). This patent covers the Zika virus strain DN-1 used in Figure 5. The authors declare no other competing interests.
Data and code availability
This study did not generate or analyze any datasets.
References
- Arguello R.J., Combes A.J., Char R., Gigan J.P., Baaziz A.I., Bousiquot E., Camosseto V., Samad B., Tsui J., Yan P., et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 2020;32:1063–1075.e7. doi: 10.1016/j.cmet.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek S.S., Watanabe S., Chan K.R., Ong E.Z., Tan H.C., Ng W.C., Nguyen M.T.X., Gan E.S., Zhang S.L., Chan K.W.K., et al. A systematic approach to the development of a safe live attenuated Zika vaccine. Nat. Commun. 2018;9:1031. doi: 10.1038/s41467-018-03337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low G.H.L., Ooi E.E., Tolfvenstam T., Leo Y.S., Hibberd M.L., Ng L.C., Lai Y.L.L., Yap G.S.L., Li C.S.C., Vasudevan S.G., et al. Early Dengue infection and outcome study (EDEN) - study design and preliminary findings. Ann. Acad. Med. Singap. 2006;35:783–789. [PubMed] [Google Scholar]
- van der Windt G.J.W., Chang C.H., Pearce E.L. Measuring bioenergetics in T cells using a seahorse extracellular flux analyzer. Curr. Protoc. Immunol. 2016;113 doi: 10.1002/0471142735.im0316bs113. 3.16B.1–3.16B.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau C., Low J.Z.H., Gan E.S., Kwek S.S., Cui L., Tan H.C., Mok D.Z.L., Chan C.Y.Y., Sessions O.M., Watanabe S., et al. Dysregulated metabolism underpins Zika-virus-infection-associated impairment in fetal development. Cell Rep. 2021;37:110118. doi: 10.1016/j.celrep.2021.110118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze any datasets.