Figure 3. Transcriptome profiling in Myocardial Infarction-Associated Transcript (MIAT) knockout (KO) mice identifies novel target genes of cardiac MIAT.
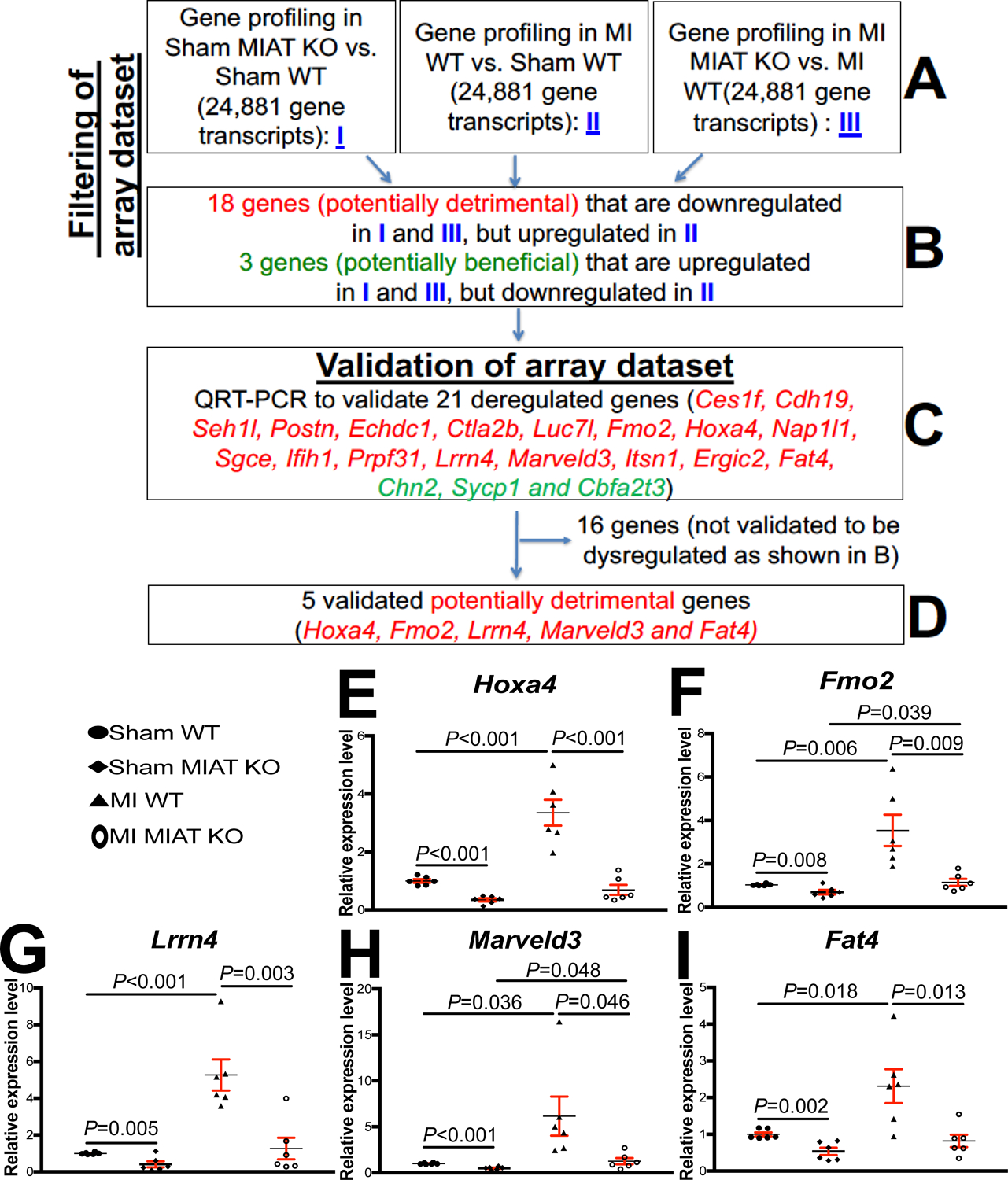
A-B, Filtering strategy of array dataset based on the correlation between genotypes and transcript signatures (I) or between cardiac phenotypes and transcript signatures (II and III). Eighteen genes, which are downregulated in both I (sham MIAT KO compared to sham wild type [WT] controls) and III (myocardial infarction [MI] MIAT KO compared to MI WT) but are upregulated in II (MI WT compared to Sham WT) at 4 weeks post-MI, were chosen for further analyses. Three genes, which are upregulated in both I (sham MIAT KO compared to sham WT controls) and III (MI MIAT KO compared to MI WT) but are downregulated in II (MI WT compared to Sham WT) at 4 weeks post-MI, were also chosen for further analyses. C-D, Validation strategy of array dataset. Twenty-one dysregulated (DE) genes were validated by Real-Time Quantitative Reverse Transcription (QRT)-PCR analyses. Note that other sixteen genes are not validated to be dysregulated as shown B. E-I, The expression of five potentially detrimental genes [homeobox a4 (Hoxa4: E), flavin-containing dimethylaniline monoxygenase 2 (Fmo2: F), leucine-rich repeat neuronal 4 (Lrrn4: G), MARVEL domain containing 3 (Marveld3: H) and FAT atypical cadherin 4 (Fat4: I)] in left ventricles (LVs) from WT and MIAT KO mice at 4 weeks post-MI. Data are shown as fold induction of gene expression normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh). N=6 per group. Two-way ANOVA with Tukey multiple comparison test.
