Figure 1. E. coli symE genetic architecture and SymE protein structural homology.
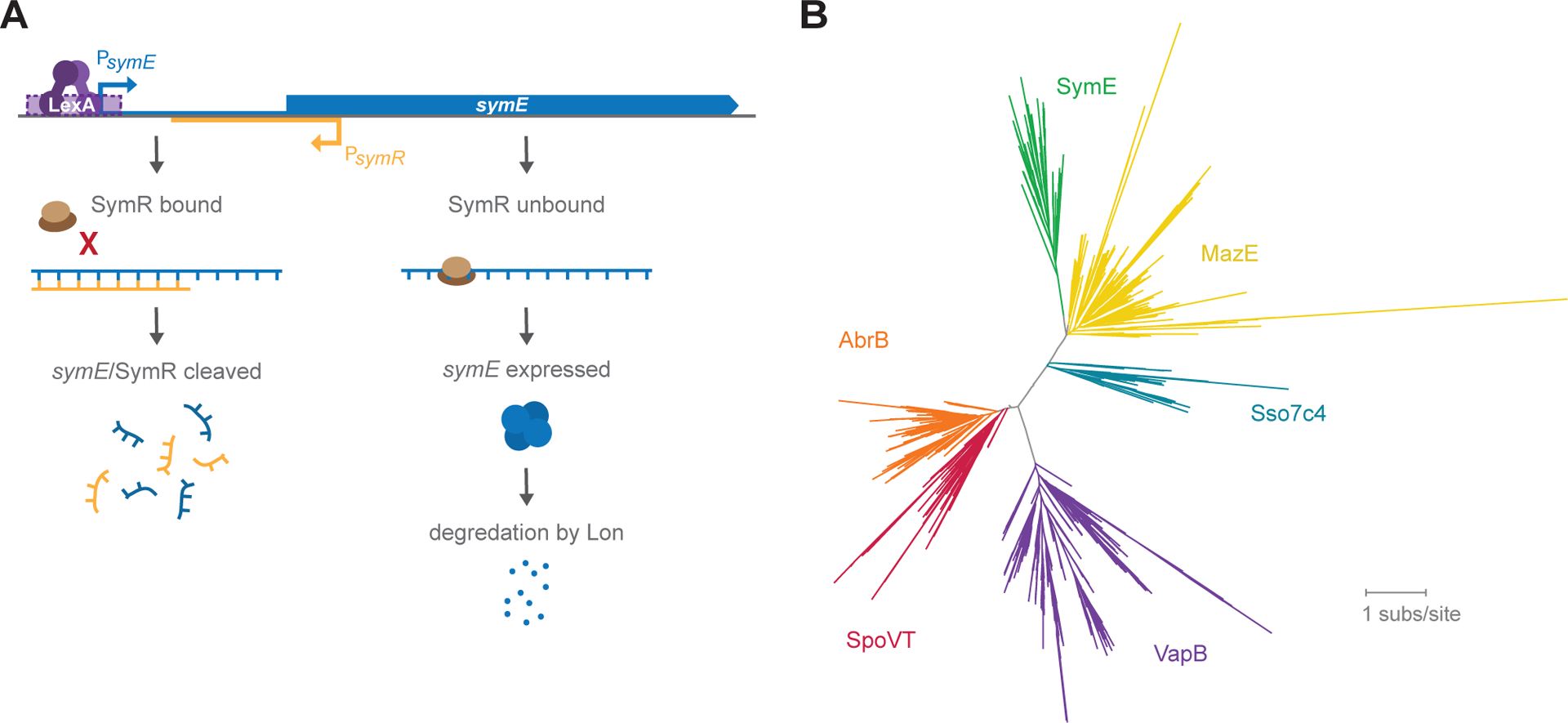
(A) symR is encoded antisense to symE and overlapping the ribosome binding site of symE. In the presence of SymR, ribosome binding to symE mRNA is occluded, and the symE/SymR complex is degraded. When there is more symE mRNA than SymR present, SymE protein is produced, but also degraded by Lon.
(B) Protein homology tree based on primary sequence alignments to SymE, indicating that SymE is likely to be most structurally similar to MazE, Sso7c4, VapB, SpoVT and AbrB.
