Figure 1.
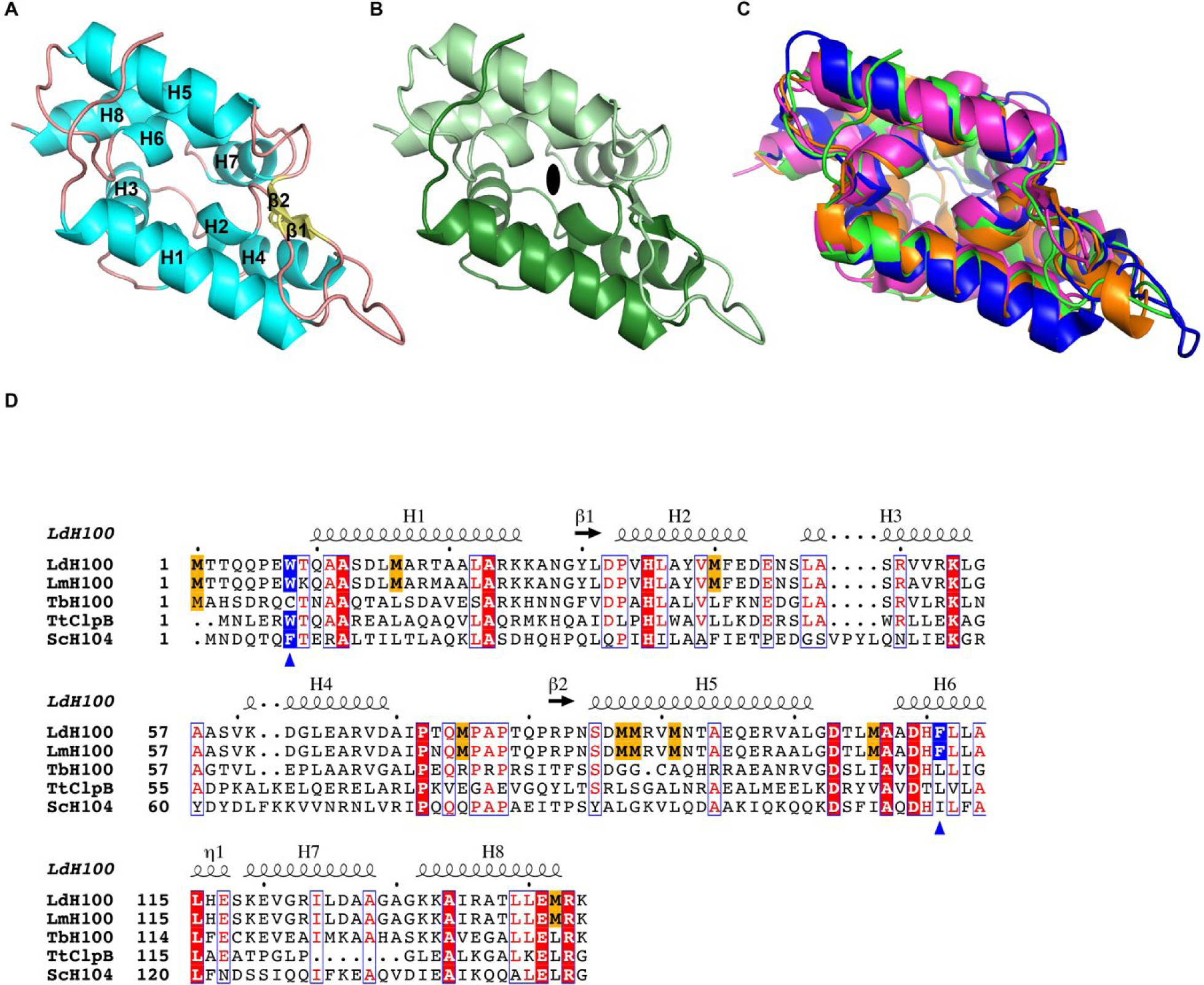
Crystal structure of the L. mexicana Hsp100N. (A) Atomic structure of LmHsp100N shown as ribbon diagram with α-helices (H1-H8) and β-strands (β1-β2) labelled sequentially. (B) View along the pseudo two-fold symmetry axis (symbol) with each Clp repeat subdomain shown in different hues. (C) Superposition of the N-domain from L. mexicana Hsp100 (green), E. coli ClpA (PDB: 1K6K; blue), T. thermophilus ClpB (PDB: 1QVR_A; orange), and B. subtilis ClpC (PDB: 2Y1Q; magenta). (D) Structural elements and sequence alignment of Hsp100 N-domains from L. donovani (LdH100), L. major (LmH100), T. brucei (TbH100), T. thermophilus (TtClpB), and S. cerevisiae (ScH104). The amino acid sequence for Hsp100N from L. mexicana (New World species) and L. donovani (Old World species) are identical. Conserved methionines in Leishmania spp. Hsp100 are highlighted in gold. The blue triangles mark the critical aromatic/hydrophobic amino acid. The dot above the sequence marks every tenth residue.
