Figure 1.
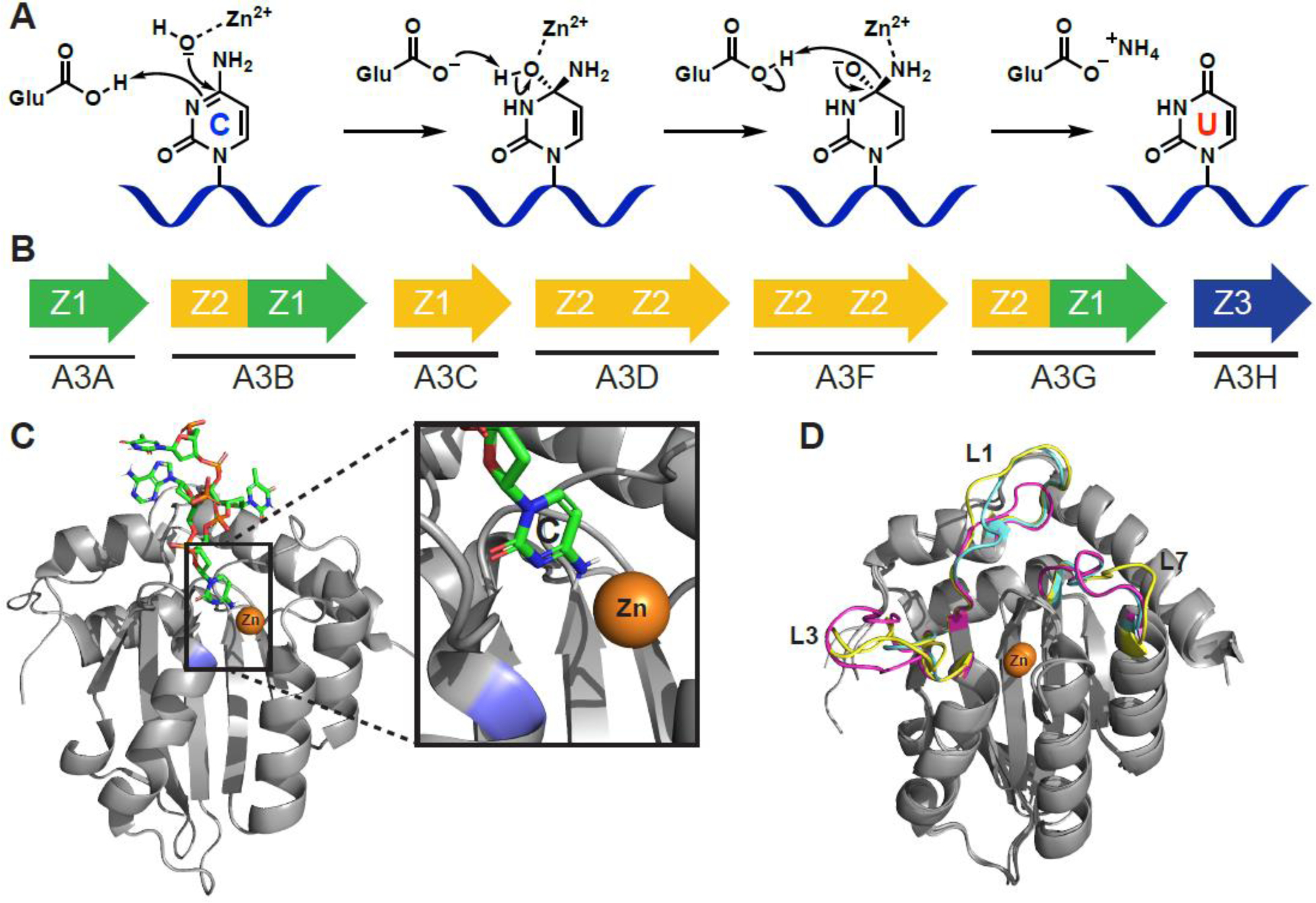
APOBEC3 structure and function. A) Proposed C-to-U hydrolytic deamination mechanism. B) The human APOBEC3 subfamily represented by arrows (single or double domain), and colors indicating phylogenetic grouping (single or double domain). C) A3B C-terminal domain co-crystal structure bound to ssDNA (PDB: 5TD5). The target cytosine is shown (inset) to be in proximity to the catalytic zinc (orange) and catalytic Glu255 (blue; mutated to Ala for crystallographic studies). D) Overlay of A3A (magenta, PDB: 4XXO), A3B C-terminal domain (cyan, PDB: 5CQI), and A3G C-terminal domain (yellow, PDB: 3IR2) showing conservationed structure between Z1 domains of different APOBEC3 enzymes of therapeutic relevance. Most of the structural variability occurs in structure is in the flexible loop regions that have significant effects on substrate binding and catalytic activity. Structure alignments performed with Pymol 2.3 align function.
