Figure 3.
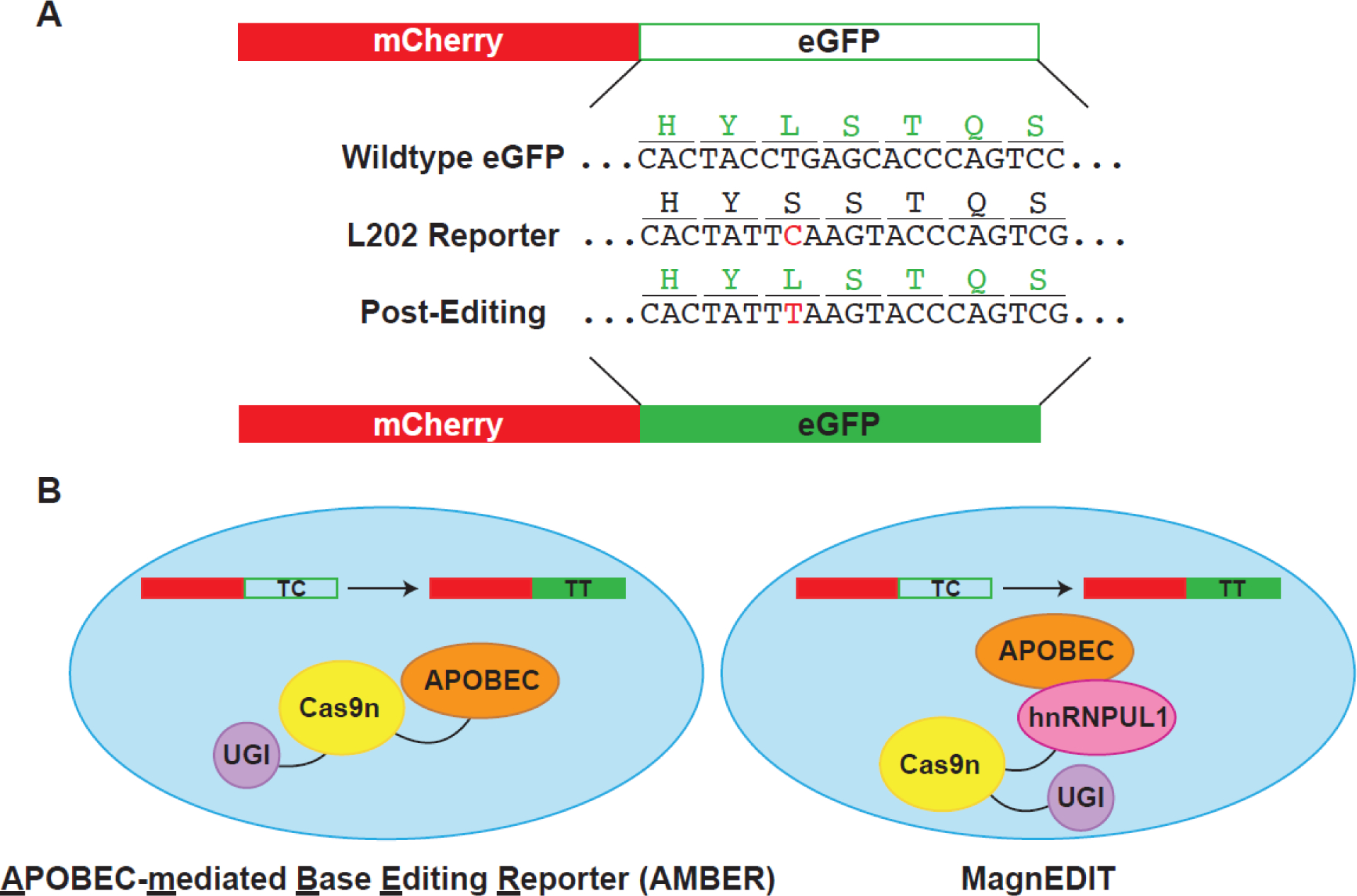
Overview of cellular assays for in vivo activity assessment. A) Plasmid editing by APOBEC-mediated Base Editing Reporter (AMBER). Reporter plasmid contains constitutively-expressed active mCherry for transfection/transduction normalization. eGFP reporter protein is inactivated by mutation of the L202 codon to encode for a serine residue, which ablates fluorescence. Silent mutations were also introduced near the editing site to reduce the opportunity for double-stranded breaks. APOBEC3 editing restores eGFP fluorescence. B) Comparison of AMBER editing system and MagnEDIT. AMBER fuses all three proteins – APOBEC3, Cas9n, and UGI – to direct editing to the eGFP reporter construct in panel A. MagnEDIT fuses Cas9n/UGI with an APOBEC-interacting protein, hnRNPUL1. gRNA directs hnRNPUL1/Cas9n/UGI to the eGFP reporter construct from panel A. hnRNPUL1 therefore acts as a “magnet” to attract the APOBEC3 protein to the reporter and restore eGFP fluorescence. Elimination of the tethering of the APOBEC3 protein to the Cas9n/UGI construct results in less off-target editing.
