Figure 1. Structure of the catalytic core of human PTase α2β2.
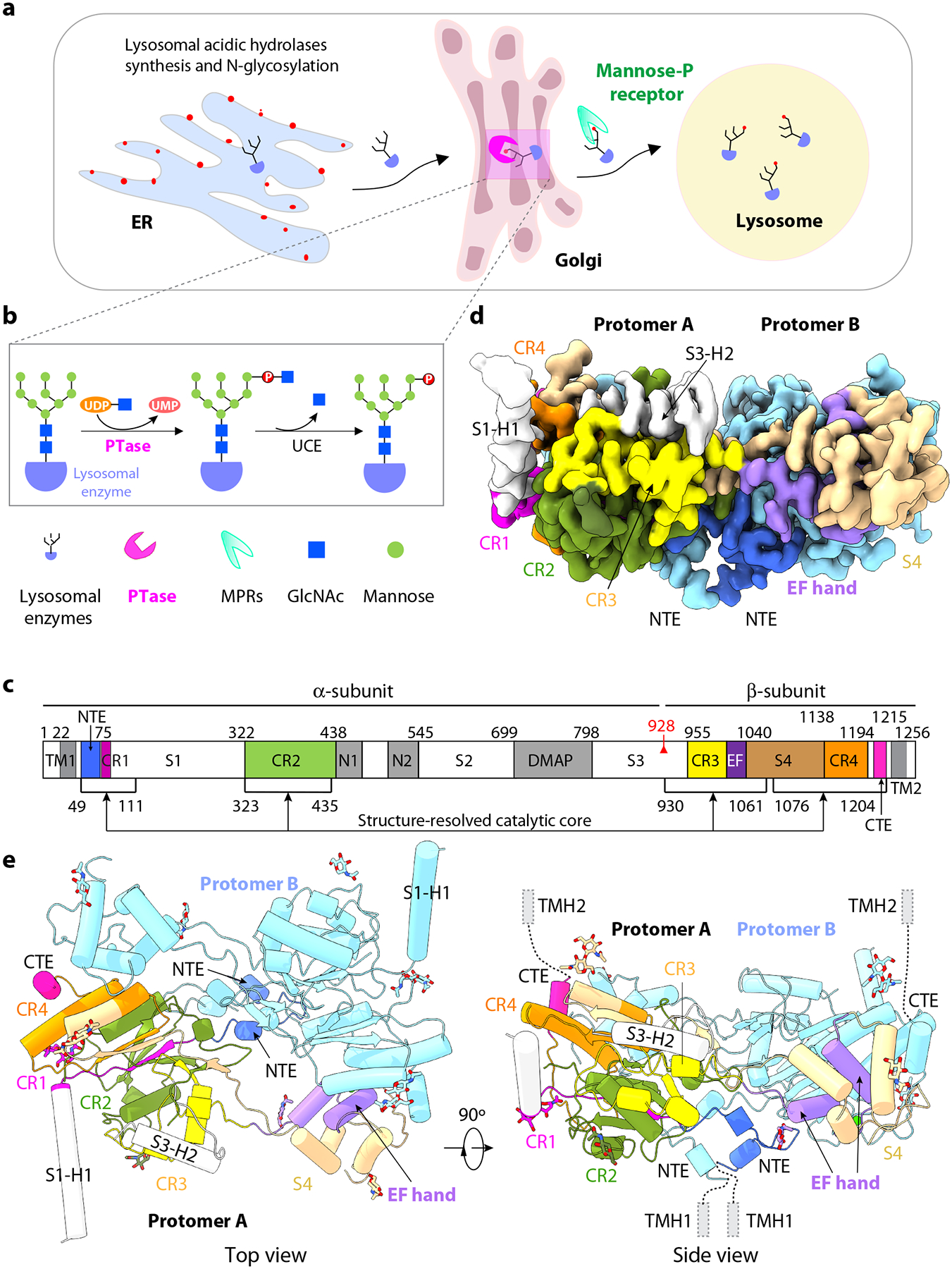
a) Schematic of the M6P-dependent targeting pathway of lysosomal hydrolases. Lysosomal hydrolases are synthesized in the ER, then transported to the Golgi where they are tagged with the M6P moiety in two steps. b) First, PTase transfers GlcNAc-1-P from UDP-GlcNAc to mannose residues of the glycan chains, then GlcNAc is removed by UCE. Lysosomal hydrolases with the M6P modification are recognized by the MPRs and transported to lysosome. c) Domain organization of human PTase. TM, transmembrane domain; CR1-4, Conserved Stealth region 1–4; S1–4, spacer regions 1–4, N1–2, Notch repeat domains 1 and 2; DMAP, DNA methyltransferase-associated protein interaction domain; EF, EF hand domain; red triangle, S1P cleavage site; Black lines, regions within atomic model. d) Cryo-EM 3D map in a side view. e) Cartoon of the atomic model of PTase core. Subunit A domains are shown in colors that match 1c. NTE, CR1-4, EF hand, S4 and C-terminus of subunit A are shown in color. Two α-helices from S1 and S3 regions are in gray. Subunit B is shown in pale cyan. The 14 resolved GlcNAc moieties at 10 glycosylation sites (5 in each subunit) are shown as sticks.
