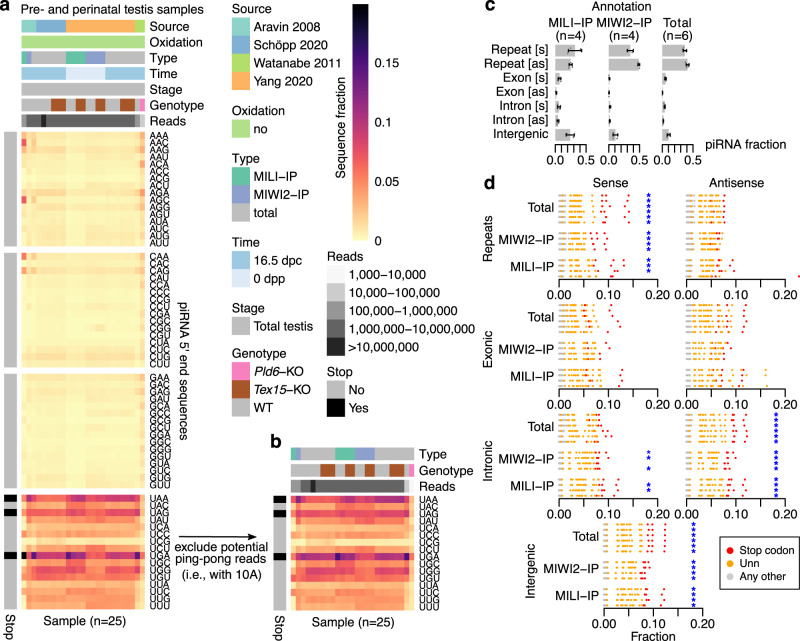
Fig. 2. Pre- and perinatal piRNAs are enriched for stop codons at their 5′ ends.
a Heatmap showing sequence distribution at piRNA 5′ ends across 25 small RNA-seq libraries from pre- and perinatal mouse testis. Each column represents one library. Column-wise annotations describe data source publication (Source), whether oxidated RNAs were captured (Oxidation), library type (Type), developmental time point (Time), spermatogenesis stage (Stage), mouse genotype (Genotype), and number of reads (Reads). Row-wise annotations show whether a sequence is a stop codon (Stop). Abbreviations: dpc, days post coitum; dpp, days postpartum; WT, wild-type. b Heatmap showing re-processing of (a) using only reads without an A at position 10. c Annotation of piRNAs across 14 samples from Yang et al.36 shown in (a). Bars represent mean fraction ± one standard deviation (sd) (4–6 replicates). Individual data points are shown in dark grey. Abbreviations: s, sense; as, antisense. d Overview of 5′ end sequences across the samples in (c) per annotation type. Each row represents one library (either WT or Tex15-KO). All possible 5′ end sequences are shown as circles and colour-coded to identify stop codons (n = 3, red), other Unn sequences (n = 13, orange), and all other sequences (n = 48, grey). Libraries where the three stop codons are more abundant than any other sequence are marked with a blue asterisk. Source data are provided as a Source Data file.