Fig. 3.
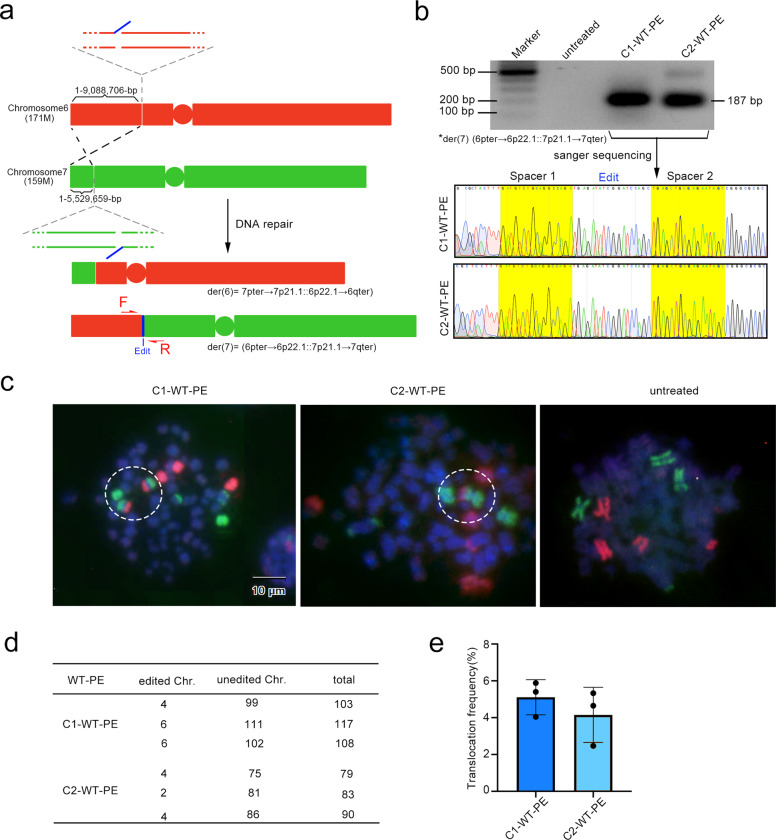
Targeted inter-chromosomal translocation by WT-PE. a Diagram showing the design of translocation between chromosome 6 and 7 pegRNAs were designed to cut a 171 Mbs fragment from the short arm of chromosome 6 and a 159 Mbs from the short arm of Chromosome 7. The edits that were complementary to each were installed on the short arm of chromosome 6 and the large fragment of chromosome 7 respectively, which we expected to induce translocations of (7pter→7p21.1::6p22.1→6qter) and (6pter→6p22.1::7p21.1→7qter). b The presence of the translocation of der (7) (6pter→6p22.1::7p21.1→7qter) was detected by PCR analysis with primers flanking each side of the translocation. Upper panel showed the agarose gel image of the resulting amplicons and lower panel showed their Sanger sequencing chromatograms with residue spacer sequences marked with yellow. However, der (6) (7pter→7p21.1::6p22.1→6qter) was not detected even with extensive PCR analysis. Primers for PCR analysis were listed in Supplementary Table 4. c Detecting the targeted translocation via FISH on metaphase cells. Two sets of probes covering the entire chromosome 6 (red) and 7 (green) were used for the FISH. Positive signals for der (7) (6pter→6p22.1::7p21.1→7qter) were observed highlighted in white dotted circles. No signals for der (6) (7pter→7p21.1::6p22.1→6qter) were observed. d, e Quantifications of the targeted translocations. d A table summarizing the translocation events that was manually counted. e Showed percentiles of the translocation