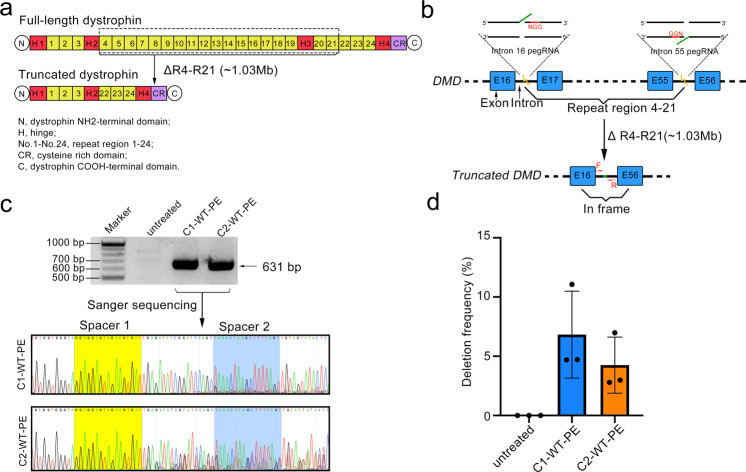
Fig. 4.
Targeted deletion of Exons 17-55 of the human DMD gene. a Diagram showing the organization of the full-length or truncated dystrophin proteins. Note that this truncated version of dystrophin has been demonstrated to be functional (ref. 9). Key domains of dystrophin protein were shown below the diagram. Dotted box highlighted the domains to be deleted. b Diagram showing the design of pegRNAs for the targeted deletion (1.03 Mbs). ssDNAs extended by WT-PE were shown in green. c The presence of the targeted deletion was detected by PCR analysis with primers flanking each side of the deletion shown in (b). Upper panel showed the agarose gel image of the resulting amplicons and lower panel showed their Sanger sequencing chromatograms with residue spacer sequences marked with yellow. Primers for PCR analysis were listed in Supplementary Table 4. d Quantifying the frequencies of targeted deletions by absolute quantitative PCR. The standard curves of wildtype- or edited-chromosome specific fragment were shown in supplementary Fig. 11. Plots showed mean ± s.d. of three independent biological replicates