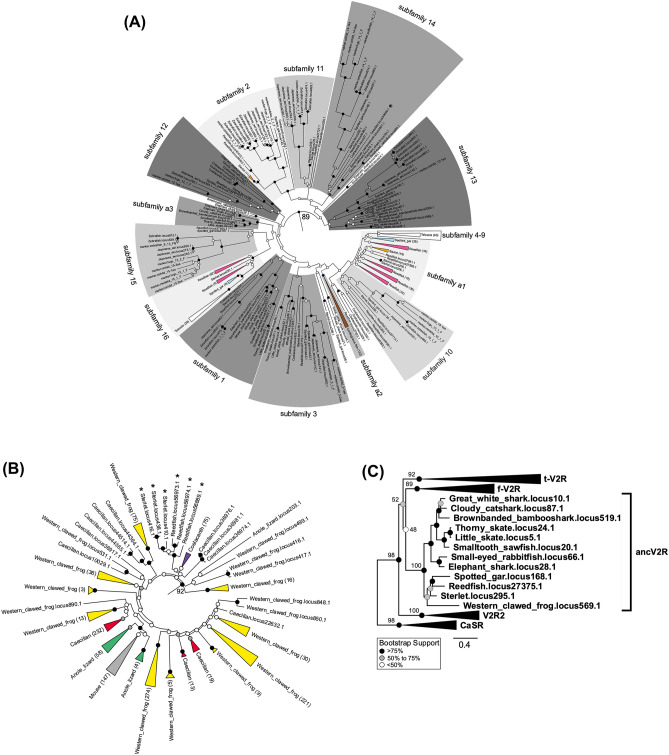
Figure 1.
Phylogenetic relationships of V2Rs of a broad range of vertebrates from cartilaginous fish to tetrapods constructed using RAxML-NG v.1.0.1 (https://github.com/amkozlov/raxml-ng). (A) Phylogenetic tree of f-V2Rs. Note that f-V2Rs were subdivided into 16 known and 3 novel subfamilies, as indicated by gray thick bars. Triangles in red, orange, blue, white, and brown indicate the expanded V2R clusters specific to reedfish, sterlet, spotted gar, cartilaginous fish, and teleost fish, respectively. Only one f-V2Rs found in the caecilian species was marked using an asterisk. (B) Phylogenetic tree of t-V2Rs. Triangles in violet, pink, yellow, green, and gray indicate expanded V2R clusters specific to coelacanth, caecilian, western clawed frog, anole lizard, and mouse, respectively. Asterisks were used to mark the t-V2Rs identified in reedfish and sterlet. Note that the t-V2Rs, in contrast to the f-V2Rs, are composed of many clusters that are expanded in a species-specific manner. (C) Overview of the phylogenetic tree of all V2Rs showing novel orthologous clade ancV2R. The calcium-sensing receptor (CaSR) gene was used for outgrouping all V2Rs. The OTU names consist of the common name and locus as summarized in Supplementary Table S1. The f-V2Rs, t-V2Rs, and V2R2 clades were compressed into black triangles. The number on the branches indicates the bootstrap support values for particular nodes. Note that the grouping of the orthologous ancV2Rs of cartilaginous fish, basal ray-finned fish, and western clawed frog was suggested by maximum bootstrap support (100%). The numbers next to triangles indicate the copy number of V2Rs included in the clusters. The filled circles on each node indicate bootstrap supports (black > 75, 75 ≧ gray ≧ 50, 50 > white). Scale bar indicates the number of amino acid substitutions per site.