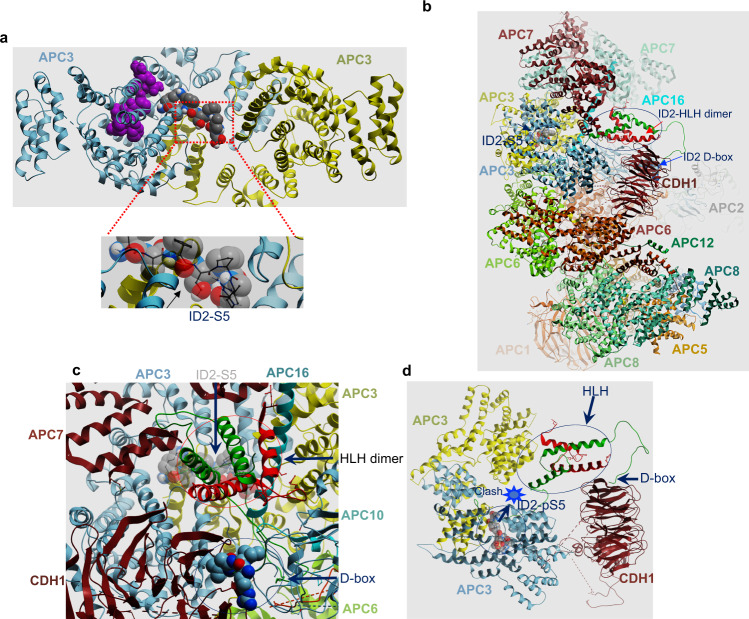
Fig. 2. Molecular docking of ID2 on the APCCDH1 complex.
a Upper panel, docked locations of ID2 amino acids 2-8 (gray-carbon, red-oxygen, blue-nitrogen, white-proton spheres) and ID2 phosphoS5 amino acids 2-8 (purple spheres) on the APC3 homodimer (gray, chain a; yellow, chain b). Lower panel, close-up view of S5 showing tight packing around the serine hydroxyl that cannot accommodate an aspartate or phosphorylation. b and c Side (b) and close-up (c) views of APCCDH1 holocomplex with ID2 in place. The APC cavity fitting ID2 bounded by APC7 (maroon helices upper left), APC16 (long aqua helix), APC10 (gray ribbon lower right) and CDH1 (maroon beta-propeller). The ID2 D-box binding peptide is shown in solid gray (carbon), red (oxygen) and blue (nitrogen) spheres, while the ID2 N-terminal peptide containing S5 docked to APC3 is shown in transparent spheres with the same color scheme. The ID2 protein is shown as green/red ribbons representing the four-helix bundle ID2 homodimer occupying the cavity with N-terminus bound to APC3, the two central helices and the C-terminal D-box bound to CDH1. APC3 homodimer is shown at the bottom of the cavity as gray and yellow ribbon. d Side view of APC3 and CDH1 only with ID2 HLH homodimer. The ID2-D-box is bound to CDH1 and the docked location of ID2-phospho-S5 amino acids 2–8 is shown as space filling spheres. Location of obstruction where the distance is too close to accommodate ID2 amino acids 9–29 is labeled “clash”.