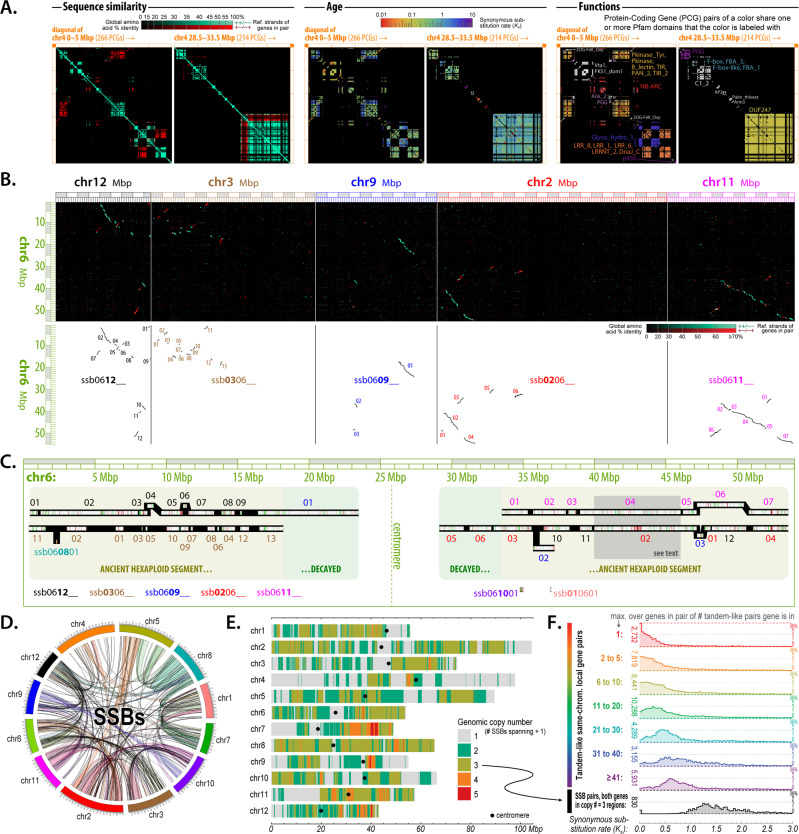
Fig. 4. Duplicated protein-coding genes.
A Sequence similarity (amino acid identity), age (synonymous substitution rate Ks), and functions (shared Pfam domains) for all pairs of proteins within two illustrative 5 Mbp regions of chr. 4. The largest block visible includes many DUF247 genes. Nearly half of Q. lobata PCGs are involved in tandem-like blocks of varying sizes (up to Mbp scales and dozens of genes at a time), often locally rearranged, and originating and growing at a variety of ages. Genes involved are diverse but enriched in certain functions. B, C With no recent whole-genome polyploidization, most of the detected PCG syntenies of Q. lobata to itself (SSBs) are small and diffuse and reflect the core eudicot triplication event γ over 100 Mya. Despite its age, this event remains quite evident—albeit highly fragmented, dispersed, and partially decayed. The whole of chr. 6 vs. the whole of chr. 12/3/9/2/11 are shown as exemplary. D, E SSBs [even without chaining as in C] cover much of the chromosomes. The highest fraction (34% of base pairs) is spanned by manifest triplication, 27% by duplication (while some duplication is recent, most appears to be decayed triplication), and 34% by no detected extant synteny. F The pairwise synonymous substitution rate (Ks) tends to be very low for genes tandemly duplicated just once (red) and increases as tandem-like block size increases (orange to violet), suggesting larger blocks are older. Ks is essentially always extremely high (≥∼1.0) for SSB gene pairs where both pair genes lie in chromosomal regions spanned by exactly two SSBs (black), supporting the syntenic triplications to be of ancient origin.