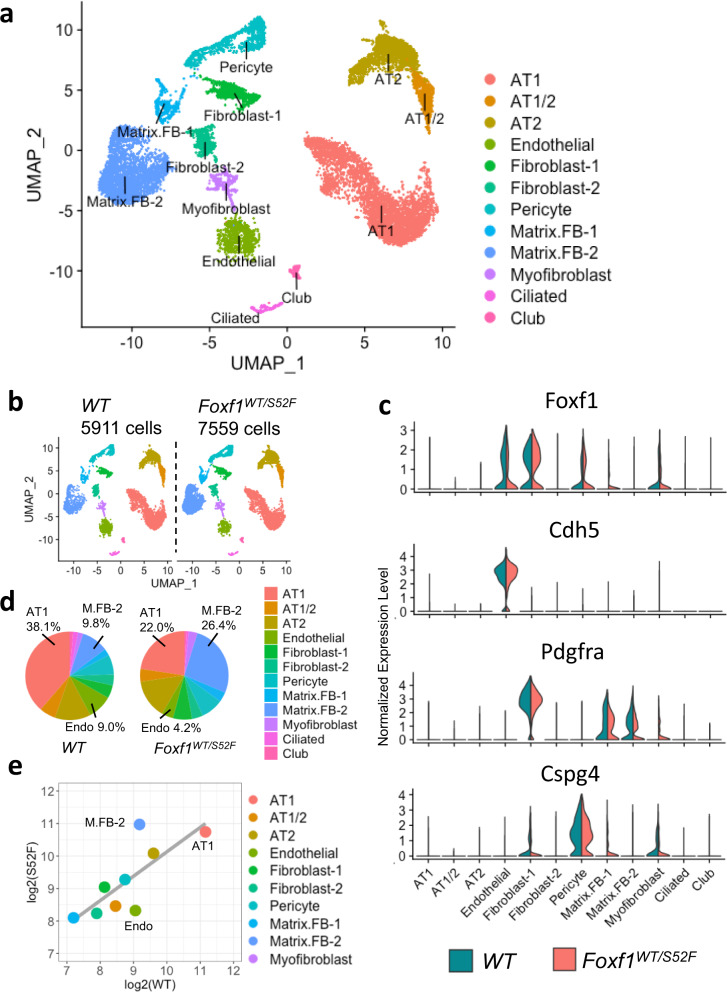
Fig. 1. Single cell RNAseq analysis identifies differences in cellular composition of the Foxf1WT/S52F lung.
a The integrated projection of CD45-depleted pulmonary cells from WT and Foxf1WT/S52F lungs. Lungs were harvested from E18.5 embryos and enzymatically digested to obtain single cell suspensions. CD45-positive cells were depleted using immunomagnetic beads. Three embryos from each genotype were pooled together prior to the scRNAseq analysis. 12 cell clusters were identified using the Uniform Manifold Approximation and Projection (UMAP) clustering. b A K-nearest neighbor (KNN) graph-based clustering approach was used to identify similar cell clusters in WT (n = 5911 cells) and Foxf1WT/S52F lungs (n = 7559 cells). c Violin plots show the presence of Foxf1 mRNA in endothelial cells, fibroblast-1, pericytes and myofibroblasts based on expression of selective cell markers Cdh5, Pdgfra and Cspg4. d Proportions of cells in each of the 12 clusters are compared in WT and Foxf1WT/S52F lungs. Percentages of endothelial and AT1 cells are decreased in Foxf1WT/S52F lungs. The percentage of matrix FB2 is increased in Foxf1WT/S52F lungs compared to lungs of WT littermates. e Linear regression analysis shows changes in distribution of endothelial, AT1 and matrix FB2 clusters between WT and Foxf1WT/S52F lungs.